Expanded genetic screening in Caenorhabditis elegans identifies new regulators and an inhibitory role for NAD+ in axon regeneration
Figures
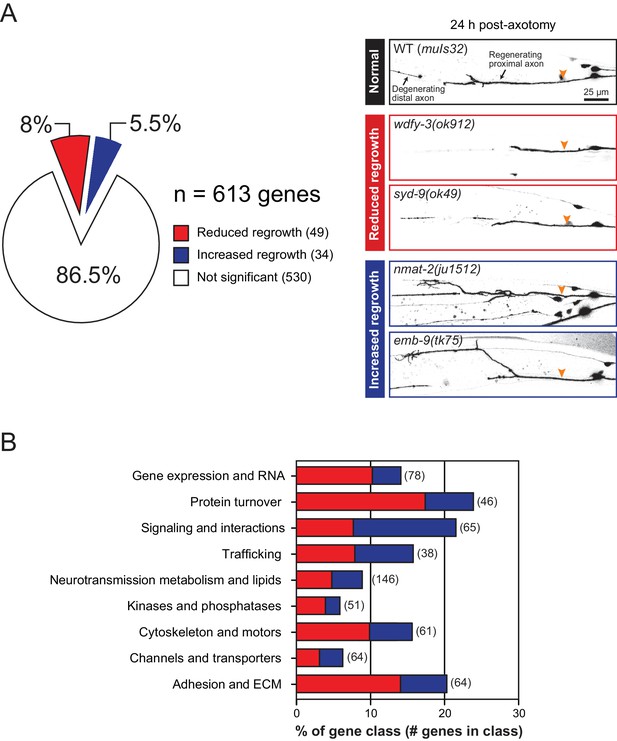
Overview and results of expanded axon regrowth screen.
(A) Pie chart showing fraction of genes screened displaying significantly reduced or increased regrowth at 24 hr. Right: representative inverted grayscale images of PLM 24 hr post-axotomy in wild type (black box), and mutants with reduced (red boxes) or increased regrowth (blue boxes). Orange arrowhead, site of axotomy. (B) Distribution of reduced/increased regrowth mutants among nine functional or structural gene classes, shown as percentage of genes in each class. See Figure 1—source data 1 for lists of genes in each class.
-
Figure 1—source data 1
List of screened genes, reference alleles, and the functional categories.
- https://doi.org/10.7554/eLife.39756.006
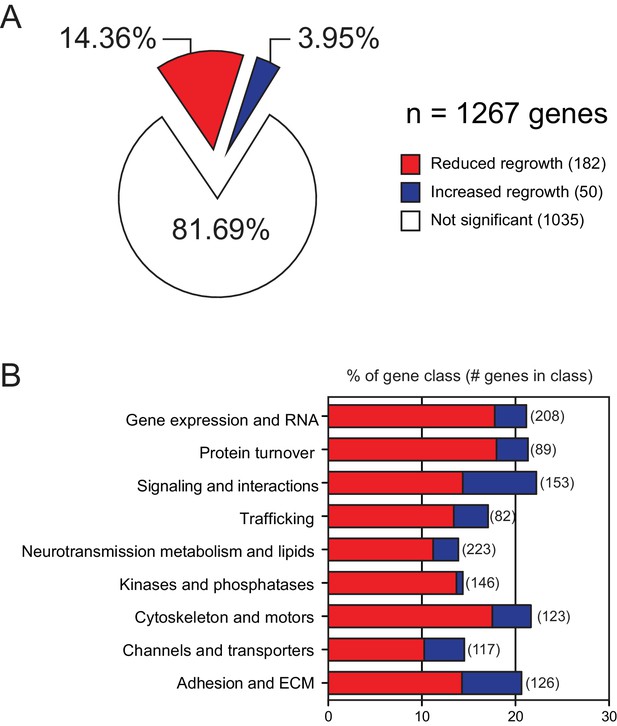
The overview and results of axon regrowth screen combined with our previous study.
(A) Pie chart showing combined genes from both screens (Chen et al., 2011)(this work) displaying significantly reduced or increased regrowth at 24 hr. (B) Distribution of reduced/increased regrowth mutants among nine functional or structural gene classes, shown as percentage of genes in each class.
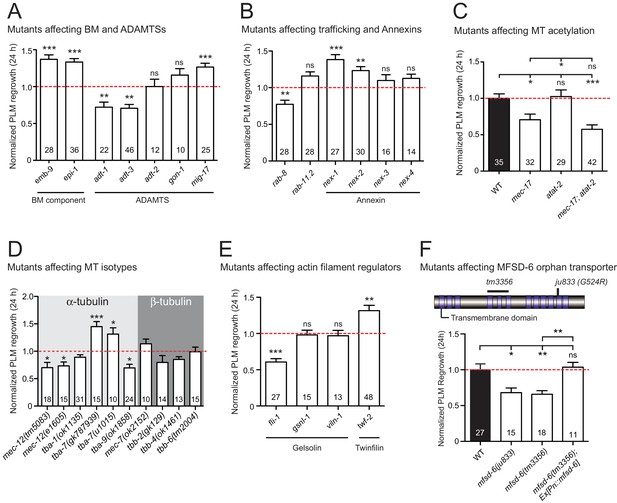
Mutants affecting multiple biological processes required for normal axon regrowth.
(A–E) Normalized PLM axon regrowth 24 hr post-axotomy in mutants affecting: (A) components of basement membrane (BM) and ADAMTS, (B) components of trafficking and Annexins, (C) microtubule acetyltransferases, (D) microtubule isotypes, and (E) actin filament regulators. Statistics, Student’s t-test with same day controls. (F) Normalized PLM axon regrowth 24 hr post-axotomy in mutants affecting mfsd-6 mutants. Pan-neuronal expression of MFSD-6 [Pn::mfsd-6] in mfsd-6(tm3356) animals rescues axon regeneration. Statistics, one-way ANOVA followed by Tukey’s multiple comparison tests. Left: MFSD-6 protein structure. Loss-of-function alleles are indicated above (tm3356, deletion; ju833, missense point mutation). Blue rectangles represent transmembrane domains. Data are shown as mean ±SEM. n, number of animals shown within columns. ns, not significant. *p<0.05; **p<0.01; ***p<0.001. In Figure 1—figure supplement 2D, tba-1, tbb-4, and tbb-6 (Chen et al., 2011) are included for comparison.
-
Figure 1—figure supplement 2—source data 1
Each data point in all graphs.
- https://doi.org/10.7554/eLife.39756.005
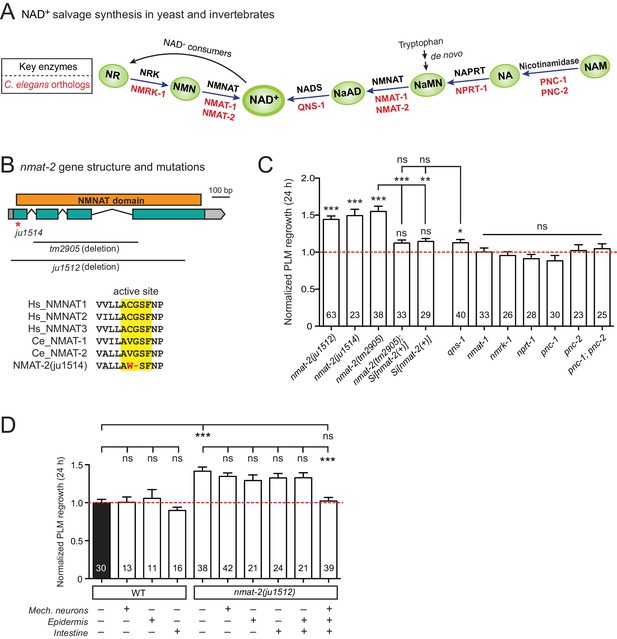
NMNAT/NMAT-2 inhibits PLM axon regrowth via its catalytic domain.
(A) Overview of NAD+ salvage biosynthesis pathway. Top, key enzymes; Bottom, C. elegans orthologs (Shaye and Greenwald, 2011). (B) Top, nmat-2 gene structure and mutant alleles. NMAT-2 contains an NMNAT domain. nmat-2(ju1514) point mutation and nmat-2(ju1512) deletion alleles were generated using CRISPR-Cas9 genome editing. Bottom, sequence alignment of the active site of NMNAT domain of C. elegans NMAT-2 (accession number: NP_492480.1; amino acids 4–14) with human NMNAT1–3 (NP_073624.2, NP_055854, NP_001307441) and C. elegans NMAT-1 (NP_510010.2). Sequences were analyzed using Clustal Omega. (C) Normalized regrowth 24 hr post-axotomy in mutants lacking genes encoding enzymes in the NAD+ biosynthesis pathway. Statistics, Student’s t-test with same day controls. For the statistical test of transgene analysis, one-way ANOVA followed by Tukey’s multiple comparison test. (D) PLM axon regrowth 24 hr post-axotomy in transgenic animals expressing nmat-2(+) driven by tissue-specific promoters for mechanosensory neurons (Pmec-4), epidermis (Pcol-12) or intestine (Pmtl-2) in a nmat-2(ju1512) background. One-way ANOVA followed by Tukey’s multiple comparison test. Data are shown as mean ± SEM. n, number of animals shown within columns. ns, not significant; *p<0.05; **p<0.01; ***p<0.001.
-
Figure 2—source data 1
Each data point in Figure 2C,D.
- https://doi.org/10.7554/eLife.39756.011
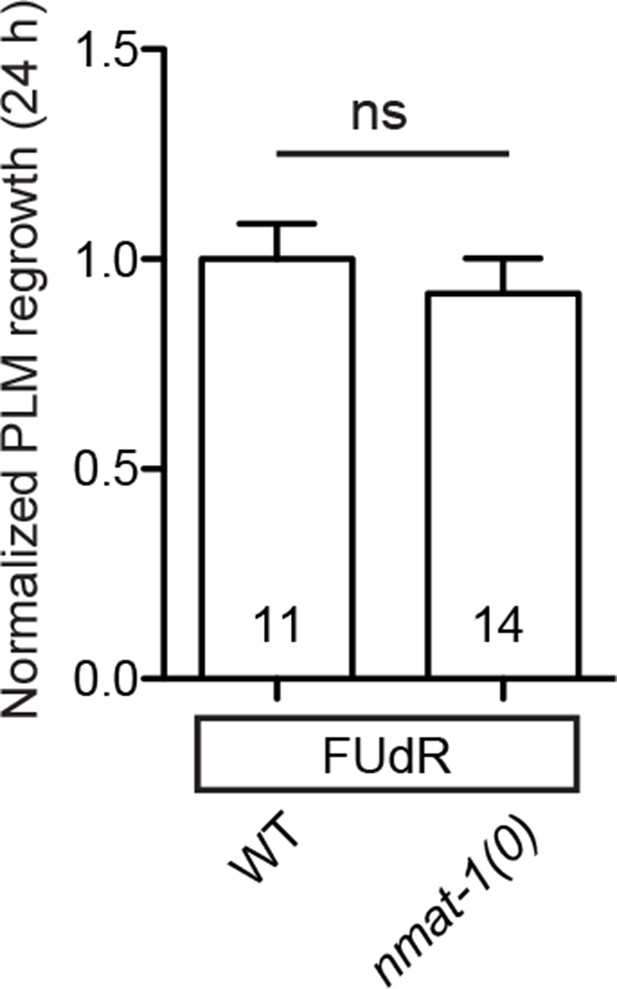
nmat-1 show no defect in PLM axon regrowth even when its germline is defective.
Normalized PLM axon regrowth 24 hr post-axotomy in mutants affecting nmat-1 mutants when treated with 50 µg/ml FUdR. Data are shown as mean ± SEM. n, number of animals shown within columns. ns, not significant.
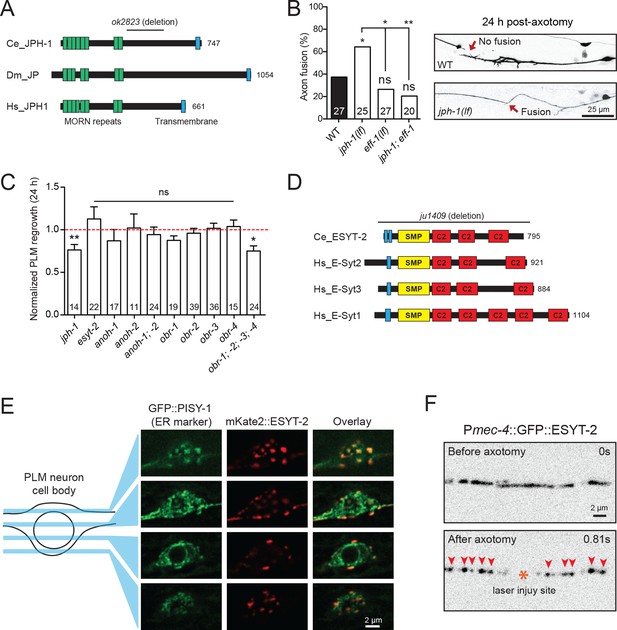
Select ER-PM membrane contact site proteins are required for axon regeneration and are sensitive to injury.
(A) Junctophilin-1 protein structure. From top to bottom: C. elegans JPH-1 (NP_492193.2), its Drosophila ortholog JP (NP_523525.2), and human ortholog JPH1 (NP_065698.1). Junctophilins contain N-terminal MORN (Membrane Occupation and Recognition Nexus) repeats (green) and a C-terminal transmembrane domain (blue). C. elegans deletion allele is indicated above (ok2823). (B) Percentage of axons that exhibit fusion between the regrowing axon and distal fragment 24 hr post-axotomy. Upper image shows a regrowing axon that has not fused with the distal fragment in a wild-type animal. Lower image shows fusion between the regrowing axon and the distal fragment in a jph-1(ok2823) animal. Fisher’s exact test. *p<0.05; **p<0.01. (C) Normalized regrowth 24 hr post-axotomy in mutants lacking selected genes encoding ER-PM MCS proteins. Data are shown as mean ± SEM. n, number of animals shown within columns. Student’s t-test with same day controls. ns, not significant; *p<0.05; **p<0.01. (D) E-Syt protein structure. From top to bottom: C. elegans ESYT-2 and its human orthologs E-Syt2, E-Syt3, and E-Syt1 (NP_065779.1, NP_114119.2, NP_056107.1, respectively). Amino acid length is indicated to the right of each protein. E-Syt proteins contain N-terminal hydrophobic regions (blue), SMP (Synaptotagmin-like Mitochondrial and lipid-binding Protein) domains (yellow), and C-terminal C2 domains (red). C. elegans deletion allele is indicated above (ju1409). (E) Images of the PLM cell body and surrounding neurites. Left, GFP::PISY-1 ER marker; Middle, mKate2::ESYT-2 driven by the mec-4 promoter; Right, Image overlays. Images show single slices taken at 1 μm intervals. (F) Representative inverted grayscale images of GFP::ESYT-2 in the axon of the PLM neuron before and immediately after axotomy (upper and lower panels, respectively). Site of laser axotomy indicated by asterisk; puncta indicated by arrowheads.
-
Figure 3—source data 1
Each data point in Figure 3C.
- https://doi.org/10.7554/eLife.39756.014
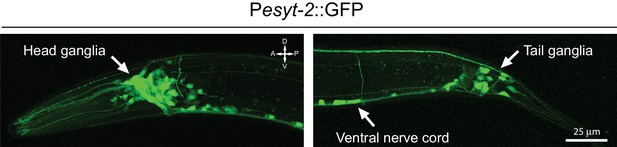
esyt-2 is widely expressed in the nervous system.
Confocal images of Pesyt-2::GFP transcriptional reporter showing widespread expression in the nervous system of L4 stage animals, including in head ganglia, ventral nerve cord, and tail ganglia. Maximum intensity projection.
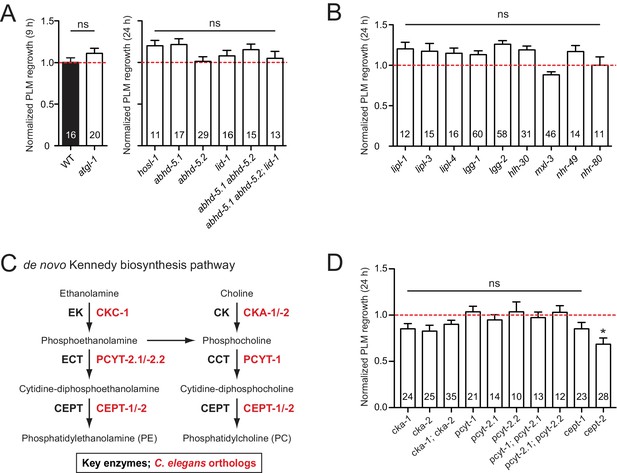
PLM axon regeneration involves membrane lipid biosynthesis pathway.
(A) Normalized PLM axon regrowth 24 hr post-axotomy in mutants affecting neutral lipolysis. (B) Normalized PLM axon regrowth 24 hr post-axotomy in mutants affecting acid lipolysis. (C) Overview of C.elegans Kennedy pathway for de novo biosynthesis of PE and PC, the major phospholipids in the PM. (D) Normalized PLM axon regrowth 24 hr post-axotomy in mutants lacking select genes encoding enzymes in the Kennedy pathway. Data are shown as mean ±SEM. n, number of animals shown within columns. Student’s t-test with same day controls. ns, not significant; *p<0.05.
-
Figure 4—source data 1
Each data point in Figure 4A,B,D.
- https://doi.org/10.7554/eLife.39756.016
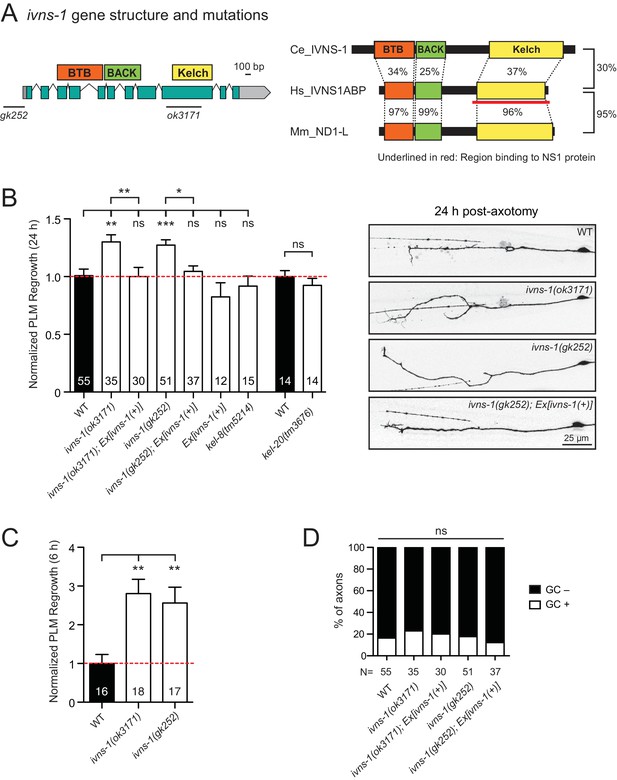
The Kelch-domain protein IVNS-1 inhibits axon regeneration.
(A) ivns-1 gene structure. Left: Loss-of-function alleles are indicated below (gk252 and ok3171). Right: Alignment of the C. elegans IVNS-1 (NP_510109.1) with its human ortholog IVNS1ABP (NP_006460.1) and mouse ortholog ND1-L (NP_473443.2). Number indicates percentage identity of protein sequences. Sequences were analyzed using Clustal Omega. (B) Normalized PLM axon regrowth 24 hr post-axotomy in mutants of Kelch-domain proteins. Data are shown as mean ±SEM. n, number of animals shown within columns. Student’s t-test with same day controls. ns, not significant; *p<0.05; **p<0.01; ***p<0.001. Right: representative inverted grayscale images of PLM 24 hr post-axotomy. Scale bar, 25 μm. (C) Normalized PLM axon regrowth 6 hr post-axotomy. Data are shown as mean ±SEM. One-way ANOVA followed by Tukey’s multiple comparison test. n, number of animals shown within columns. **p<0.01. (D) Percentage of axons with growth cones (GCs) 6 hr post-axotomy. n, Number of animals shown below columns. Fisher’s exact test. ns, not significant.
-
Figure 5—source data 1
Each data point in Figure 5B,C.
- https://doi.org/10.7554/eLife.39756.018
Tables
Mutants displaying reduced PLM regrowth
https://doi.org/10.7554/eLife.39756.007| Gene name | Mutations | Normalized regrowth (24 hr) | N | P value | Molecular function | Closest human Genea |
|---|---|---|---|---|---|---|
| A. Cell Adhesion and ECM | ||||||
| adt-1 | cn30 | 0.75 | 21 | ** | ADAM metalloprotease | ADAMTS3 |
| adt-3 | ok923 | 0.71 | 46 | ** | ADAM metalloprotease | ADAMTS2 |
| C05D9.7 | ok2931 | 0.60 | 28 | *** | Unknown | N/A |
| dpy-10 | e128 | 0.67 | 28 | ** | 5FMC ribosome biogenesis complex | PELP1 |
| F35G2.1 | ok1669 | 0.68 | 27 | *** | Quiescin sulfhydryl oxidase | QSOX1 |
| gly-2 | tm839 | 0.69 | 16 | * | Mannosyl-glycoprotein N-acetylglucosaminyltransferases | MGAT5 |
| osm-11 | rt142 | 0.72 | 13 | ** | Secreted protein | N/A |
| zig-1 | ok784 | 0.68 | 12 | * | Basigin | BSG |
| zig-3 | ok1476 | 0.73 | 18 | ** | Kazal type serine peptidase inhibitor domain | KAZALD1 |
| B. Channels and transporters | ||||||
| abts-1 | ok1566 | 0.74 | 34 | *** | Anion exchange protein | SLC4A7 |
| mfsd-6 | ju833 | 0.68 | 15 | ** | Major facilitator | MFSD6 |
| C. Cytoskeleton and motors | ||||||
| fli-1 | ky535 | 0.61 | 27 | *** | Actin remodeling protein | FLII |
| mec-12 | e1605 | 0.73 | 15 | * | Tubulin α−3 chain | TUBA1C |
| mec-17 | ok2109 | 0.55 | 14 | ** | α-Tubulin N-acetyltransferase | ATAT1 |
| tba-9 | ok1858 | 0.70 | 24 | ** | α-Tubulin | TUBA3 |
| vab-10 | e698 | 0.78 | 11 | * | Spectraplakin | DST |
| D. Protein kinases and phosphatases | ||||||
| plk-1 | or683ts | 0.59 | 13 | ** | Polo like kinase | PLK1 |
| svh-2 | tm737 | 0.68 | 30 | *** | Receptor Tyrosine kinase | MET |
| E. Neurotransmission, metabolism, and lipid | ||||||
| cept-2 | ok3135 | 0.68 | 27 | * | Choline/ethanolamine phosphotransferase | CEPT1 |
| cpr-1 | ok1344 | 0.60 | 32 | *** | Cysteine proteinase | CTSB |
| dhhc-11 | gk1105 | 0.75 | 32 | *** | Palmitoyltransferase | ZDHHC11 |
| eat-3 | tm1107 | 0.74 | 16 | ** | Mitochondrial dynamin like GTPase | OPA1 |
| npr-20 | ok2575 | 0.49 | 44 | *** | G-protein coupled receptor | CCKBR/TRHR |
| ptps-1 | tm1984 | 0.55 | 27 | *** | 6-pyruvoyl tetrahydrobiopterin synthase | PTS |
| supr-1 | ju1118 | 0.78 | 30 | ** | Unknown | N/A |
| F. Trafficking | ||||||
| jph-1 | ok2823 | 0.77 | 14 | ** | Junctophilin | JPH1 |
| rab-8 | tm2526 | 0.77 | 28 | ** | Ras GTPase | RAB8B |
| rsef-1 | ok1356 | 0.66 | 17 | * | Endosomal Rab family GTPase | RASEF |
| G. Signaling and interactions | ||||||
| osm-7 | tm2256 | 0.57 | 28 | ** | Unknown | N/A |
| par-2 | or373 | 0.63 | 47 | *** | C3HC4-type RING-finger | TRIM |
| rgl-1 | ok1921 | 0.74 | 30 | ** | Ral guanine nucleotide dissociation stimulator | RGL1 |
| wdfy-3 | ok912 | 0.52 | 17 | *** | WD40 and FYVE domain | WDFY3 |
| wdr-23 | tm1817 | 0.66 | 30 | *** | DDB1 and CUL4 associated factor | DCAF11 |
| H. Protein turnover, proteases, cell death | ||||||
| brap-2 | ok1492 | 0.61 | 62 | *** | BRCA1-associated protein; zinc ion binding activity | BRAP |
| cdc-48.1 | tm544 | 0.69 | 19 | ** | Transitional ER ATPase homolog | VCP |
| ced-9 | n1950 | 0.73 | 16 | * | Cell-death inhibitor Bcl-2 homolog | BCL2 |
| dnj-23 | tm7102 | 0.69 | 32 | ** | DNaJ domain (prokaryotic heat shock protein) | DNAJC9 |
| fbxc-50 | tm5154 | 0.73 | 12 | * | F-box protein | N/A |
| math-33 | ok2974 | 0.64 | 12 | *** | Ubiquitin-specific protease | USP7 |
| skr-5 | ok3068 | 0.69 | 12 | * | S-phase kinase associated protein | SKP1 |
| tep-1 | tm3720 | 0.53 | 36 | *** | ThiolEster containing Protein; endopeptidase inhibitor activity | CD109 |
| I. Gene expression and RNA regulation | ||||||
| mec-8 | e398 | 0.23 | 16 | *** | RNA binding protein, mRNA processing factor | RBPMS |
| rict-1 | mg360 | 0.58 | 26 | *** | Subunit of TORC2 | RICTOR |
| rtcb-1 b | gk451 | 0.58 | 25 | *** | tRNA-splicing ligase RtcB homolog | RTCB |
| skn-1 | ok2315 | 0.78 | 10 | * | Basic leucine zipper protein | NFE2 |
| smg-3 | r930 | 0.68 | 28 | *** | Nonsense mediated mRNA decay regulator | UPF2 |
| syd-9 | ju49 | 0.47 | 15 | *** | Zinc finger E-box binding homeobox | ZEB1 |
| tdp-1 | ok803 | 0.70 | 36 | *** | TAR DNA-binding protein | TARDBP/TDP-43 |
| wdr-5.1 | ok1417 | 0.70 | 26 | *** | WD repeat-containing protein | WDR5 |
-
Genes are classified in nine functional or structural classes. Mutations are genetic or predicted molecular nulls, or partial loss-of-function. Normalized regrowth is relative to matched same-day controls or to pooled controls. Significant levels (*p<0.05; **p<0.01; ***p<0.001) based on Student’s t-test.
a Closest human gene based on BLASTP score in Wormbase WS263; Ensembl/HGNC symbol.
-
brtcb-1(gk451) mutant reported to show increased regrowth in the C. elegans motor neurons (Kosmaczewski et al., 2015).
Mutants displaying increased PLM regrowth
https://doi.org/10.7554/eLife.39756.008| Gene name | Mutations | Normalized regrowth (24 hr) | N | P value | Molecular function | Closest human Genea |
|---|---|---|---|---|---|---|
| A. Cell adhesion and ECM | ||||||
| emb-9 | tk75 b | 1.37 | 28 | *** | Collagen type IV α3 chain | COL4A3 |
| epi-1 | gm121 | 1.33 | 36 | *** | Laminin | LAMA |
| mig-17 | k174 | 1.24 | 37 | *** | ADAM metalloprotease | ADAMTS5 |
| ZC116.3 | ok1618 | 1.40 | 26 | *** | Cubilin | CUBN |
| B. Channels and transporters | ||||||
| lgc-12 | ok3546 | 1.33 | 26 | ** | Serotonin receptor 3E | HTR3E |
| tmc-1 | ok1859 | 1.31 | 30 | ** | Transmembrane channel-like protein | TMC1 |
| C. Cytoskeleton and motors | ||||||
| ivns-1 | ok3171 | 1.31 | 18 | ** | Actin-binding; splicing | IVNS1ABP |
| twf-2 | ok3564 | 1.33 | 39 | ** | Twinfilin actin binding protein | TWF |
| nud-1 | ok552 | 1.30 | 25 | ** | Nuclear distribution C, Dynein complex regulator | NUDC |
| tba-7 | gk787939 | 1.45 | 15 | *** | α-tubulin | TUBA |
| E. Neurotransmission, metabolism, and lipid | ||||||
| nmat-2 | tm2905 | 1.55 | 38 | *** | Nicotinamide mononucleotide adenylyltransferase | NMNAT1 |
| qns-1 | ju1563 | 1.13 | 40 | * | NAD + synthetase | NADSYN1 |
| mgl-1 | tm1811 | 1.24 | 28 | ** | Glutamate metabotropic receptor | GRM3 |
| mgl-3 | tm1766 | 1.32 | 22 | ** | Glutamate metabotropic receptor | GRM6 |
| npr-25 | ok2008 | 1.27 | 26 | ** | Coagulation factor II thrombin receptor | F2RL2 |
| ucr-2.3 | ok3073 | 1.41 | 24 | *** | Ubiquinol-cytochrome C reductase core protein | UQCRC2 |
| F. Trafficking | ||||||
| nex-1 c | gk148 | 1.38 | 27 | *** | Annexin | ANXA13 |
| nex-2 d | ok764 | 1.23 | 30 | ** | Annexin | ANXA7 |
| snb-6 | tm5195 | 1.30 | 38 | ** | Vesicle associated membrane protein | VAMP1 |
| G. Signaling and interactions | ||||||
| drag-1 | tm3773 | 1.52 | 26 | ** | Repulsive guidance molecule BMP co-receptor | RGMB |
| ect-2 | ku427 | 1.31 | 31 | ** | RhoGEF | ECT2 |
| lin-2 | e1309 | 1.31 | 27 | *** | Membrane associated guanylate kinase | CASK |
| magi-1 | zh66 | 1.40 | 29 | *** | Membrane associated guanylate kinase | MAGI2 |
| prmt-5 | gk357 | 1.26 | 24 | ** | Protein arginine N-methyltransferase | PRMT5 |
| rap-1 | tm861 | 1.33 | 10 | *** | Ras small GTPase | RAP1 |
| smz-1 | ok3576 | 1.39 | 13 | * | PDZ domain-containing protein | N/A |
| trxr-1 | tm2047 | 1.3 | 31 | * | Thioredoxin reductase | TXNRD2 |
| H. Protein turnover, proteases, cell death | ||||||
| natb-1 | ju1405 | 1.29 | 14 | * | N(α)-acetyltransferase 20 | NAA20 |
| rnf-5 | tm794 | 1.28 | 15 | * | Ring finger protein | RNF5 |
| ulp-5 | tm3063 | 1.22 | 30 | *** | SUMO specific peptidase | SENP7 |
| I. Gene expression and RNA regulation | ||||||
| csr-1 | fj54 | 1.24 | 38 | *** | Argonaute | AGO1 |
| hda-6 | tm3436 | 1.29 | 42 | *** | Histone deacetylase | HDAC6 |
| elpc-3 | ok2452 | 1.31 | 26 | *** | Elongator acetyltransferase complex subunit | ELP3 |
-
Genes are classified in nine functional or structural classes. Mutations are genetic or predicted molecular nulls, or partial loss-of-function. Normalized regrowth is relative to matched same-day controls or to pooled controls. Significant levels (*p<0.05; **p<0.01; ***p<0.001) based on Student’s t-test.
a Closest human gene based on BLASTP score in Wormbase WS263; Ensembl/HGNC symbol.
-
b emb-9(tk75) mutant reported to be a gain-of-function allele that makes stable EMB-9/Type IV collagen (Kubota et al., 2012).
cnex-1(gk148) mutant reported to show reduced regrowth in the C. elegans motor neurons (Nix et al., 2014).
-
d nex-2(bas4) mutant reported to show normal regrowth in the C. elegans motor neurons (Nix et al., 2014).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Bacterial strain | E. coli: OP50 | Caenorhabditis Genetics Center | RRID: WB-STRAIN:OP50 | |
| Genetic reagent (C. elegans) | Strain wild type N2 | Caenorhabditis Genetics Center | RRID:WB-STRAIN:N2_ (ancestral) | |
| Genetic reagent (C. elegans) | CZ10969: Pmec-7-GFP(muIs32) II | Considered as‘WT’ in many axotomy experiments | ||
| Genetic reagent (C. elegans) | CZ10175: Pmec-4-GFP(zdIs5) I | Considered as ‘WT’ in many axotomy experiments | ||
| Genetic reagent (C. elegans) | CZ25411: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ25415: nmat-2(ju1514) I/hT2 I, III; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ17633: nmat-2(tm2905) I/hT2 I, III; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ24324: Pmec-7-GFP(muIs32) II; qns-1(ju1563) IV/mIs11 sd IV | |||
| Genetic reagent (C. elegans) | CZ25642: Pmec-4-GFP(zdIs5) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C.elegans) | CZ25534: nmrk-1(ok2571) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ24241: Pmec-4-GFP(zdIs5) I; nprt-1(tm6342) IV | |||
| Genetic reagent (C. elegans) | CZ24242: Pmec-4-GFP(zdIs5) I; pnc-1(tm3502) IV | |||
| Genetic reagent (C. elegans) | CZ24802: Pmec-4-GFP(zdIs5) I; pnc-2(tm6438) IV | |||
| Genetic reagent (C. elegans) | CZ25466: Pmec-7-GFP(muIs32) II; juSi347[nmat-2 gDNA] IV | |||
| Genetic reagent (C. elegans) | CZ25469: nmat-2(tm2905) I;Pmec-7-GFP(muIs32) II; juSi347[nmat-2 gDNA] IV | |||
| Genetic reagent (C. elegans) | CZ26216: Pmec-7-GFP(muIs32) II; Ex[Pmec-4-nmat-2(juEx7834)] | |||
| Genetic reagent (C. elegans) | CZ26217: Pmec-7-GFP(muIs32) II; Ex[Pmec-4-nmat-2(juEx7835)] | |||
| Genetic reagent (C. elegans) | CZ26220: Pmec-7-GFP(muIs32) II; Ex[Pcol-12-nmat-2(juEx7838)] | |||
| Genetic reagent (C. elegans) | CZ26221: Pmec-7-GFP(muIs32) II; Ex[Pcol-12-nmat-2(juEx7839)] | |||
| Genetic reagent (C. elegans) | CZ26218: Pmec-7-GFP(muIs32) II; Ex[Pmtl-2-nmat-2(juEx7836)] | |||
| Genetic reagent (C. elegans) | CZ26219: Pmec-7-GFP(muIs32) II; Ex[Pmtl-2-nmat-2(juEx7837)] | |||
| Genetic reagent (C. elegans) | CZ26222: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmec-4-nmat-2(juEx7840)] | |||
| Genetic reagent (C. elegans) | CZ26223: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmec-4-nmat-2(juEx7841)] | |||
| Genetic reagent (C. elegans) | CZ26224: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pcol-12-nmat-2(juEx7842)] | |||
| Genetic reagent (C. elegans) | CZ26225: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pcol-12-nmat-2(juEx7843)] | |||
| Genetic reagent (C. elegans) | CZ26310: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmtl-2-nmat-2(juEx7836)] | |||
| Genetic reagent (C. elegans) | CZ26311: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmtl-2-nmat-2(juEx7837)] | |||
| Genetic reagent (C. elegans) | CZ26332: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmtl-2, col-12::nmat-2(juEx7853)] | |||
| Genetic reagent (C. elegans) | CZ26333: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmtl-2, col-12::nmat-2(juEx7854)] | |||
| Genetic reagent (C. elegans) | CZ26285: nmat-2(ju1512) I/hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmtl-2, col-12, mec-4::nmat-2(juEx7850)] | |||
| Genetic reagent (C. elegans) | CZ26286: nmat-2(ju1512) I / hT2 I, III; Pmec-7-GFP(muIs32) II; Ex[Pmtl-2, col-12, mec-4::nmat-2(juEx7851)] | |||
| Genetic reagent (C. elegans) | CZ26391: jph-1(ok2823) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ22032: Pmec-4-GFP(zdIs5) I; anoh-1(tm4762) III | |||
| Genetic reagent (C. elegans) | CZ22033: Pmec-4-GFP(zdIs5) I; anoh-2(tm4796) IV | |||
| Genetic reagent (C. elegans) | CZ26325: Pmec-4-GFP(zdIs5) I; anoh-1(tm4762) III; anoh-2(tm4796) IV | |||
| Genetic reagent (C. elegans) | CZ26069: Pmec-4-GFP(zdIs5) I; obr-1(xh16) III | |||
| Genetic reagent (C. elegans) | CZ24555: Pmec-4-GFP(zdIs5) I; obr-2 (xh17) V | |||
| Genetic reagent (C. elegans) | CZ24556: Pmec-4-GFP(zdIs5) I; obr-3(tm1087) X | |||
| Genetic reagent (C. elegans) | CZ24557: obr-4(tm1567) I; Pmec-4-GFP(zdIs5) I | |||
| Genetic reagent (C. elegans) | CZ25696: obr-4(tm1567) I; Pmec-4-GFP(zdIs5) I; obr-1(xh16) III; obr-2(xh17) V; obr-3(tm1087) X | |||
| Genetic reagent (C. elegans) | CZ26375: Pmec-4-GFP(zdIs5) I; esyt-2(ju1409) III | |||
| Genetic reagent (C. elegans) | CZ26570: juIs540 [Pmec-4-mKate2-ESYT-2]; juEx7807[Pmec-4-GFP-PISY-1] | |||
| Genetic reagent (C. elegans) | CZ24897: juEx7604 [Pmec-4-GFP-ESYT-2] | |||
| Genetic reagent (C. elegans) | CZ22087: Pmec-7-GFP(muIs32) II; atgl-1(tm3116) III / hT2 I, III | |||
| Genetic reagent (C. elegans) | CZ22536: Pmec-7-GFP(muIs32) II; hosl-1(gk278589) X | |||
| Genetic reagent (C. elegans) | CZ22006: Pmec-7-GFP(muIs32) II; abhd-5.1(ok3722) V | |||
| Genetic reagent (C. elegans) | CZ21968: Pmec-7-GFP(muIs32) II; abhd-5.2(ok3245) V | |||
| Genetic reagent (C. elegans) | CZ22007: lid-1(gk575511) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ22163: Pmec-7-GFP(muIs32) II; abhd-5.2(ok3245) V abhd-5.1(ju1282) V | |||
| Genetic reagent (C. elegans) | CZ22166: lid-1(gk575511) I; Pmec-7-GFP(muIs32) II; abhd-5.2(ok3245) abhd-5.1(ju1282) V | |||
| Genetic reagent (C. elegans) | CZ22686: Pmec-7-GFP(muIs32) II; lipl-1(tm1954) V | |||
| Genetic reagent (C. elegans) | CZ22688: Pmec-7-GFP(muIs32) II; lipl-3(tm4498) V | |||
| Genetic reagent (C. elegans) | CZ22535: Pmec-7-GFP(muIs32) II; lipl-4(tm4417) V | |||
| Genetic reagent (C. elegans) | CZ24364: Pmec-4-GFP(zdIs5) I; lgg-1(tm3489) II/ mIn1 II | |||
| Genetic reagent (C. elegans) | CZ23325: Pmec-7-GFP(muIs32) II; lgg-2(tm6474) IV | |||
| Genetic reagent (C. elegans) | CZ23322: Pmec-7-GFP(muIs32) II; hlh-30(tm1978) IV | |||
| Genetic reagent (C. elegans) | CZ14408: Pmec-7-GFP(muIs32) II; mxl-3(ok1947) X | |||
| Genetic reagent (C. elegans) | CZ22541: nhr-49(nr2041) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ22510: Pmec-7-GFP(muIs32) II; nhr-80(tm1011) III | |||
| Genetic reagent (C. elegans) | CZ25587: Pmec-4-GFP(zdIs5) I; cka-1(tm1241) IV | |||
| Genetic reagent (C. elegans) | CZ25549: Pmec-4-GFP(zdIs5) I; cka-2(tm841) X | |||
| Genetic reagent (C. elegans) | CZ25403: Pmec-4-GFP(zdIs5) I; cka-1(tm1241) IV; cka-2(tm841) X. | |||
| Genetic reagent (C. elegans) | CZ25370: Pmec-4-GFP(zdIs5) I; pcyt-1(et9) X | |||
| Genetic reagent (C. elegans) | CZ25790: Pmec-4-GFP(zdIs5) I; pcyt-2.1(gk440213) I | |||
| Genetic reagent (C. elegans) | CZ25368: Pmec-4-GFP(zdIs5) I; pcyt-2.2(ok2179) X | |||
| Genetic reagent (C. elegans) | CZ25992: Pmec-4-GFP(zdIs5) I; pcyt-2.1(gk440213) I; pcyt-2.2(ok2179) X | |||
| Genetic reagent (C. elegans) | CZ26521: Pmec-4-GFP(zdIs5) I; pcyt-2.1(gk440213) I; pcyt-1(et9) X | |||
| Genetic reagent (C. elegans) | CZ25369: Pmec-4-GFP(zdIs5) I; cept-1(et10) X | |||
| Genetic reagent (C. elegans) | CZ26423: Pmec-4-GFP(zdIs5) I; cept-2(ok3135) V | |||
| Genetic reagent (C. elegans) | CZ19835: Pmec-4-GFP(zdIs5) I; kel-8(tm5214) V | |||
| Genetic reagent (C. elegans) | CZ23911: kel-20(tm3676) I; mec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ18224: Pmec-4-GFP(zdIs5) I; ivns-1(ok3171) X | |||
| Genetic reagent (C. elegans) | CZ18225: Pmec-4-GFP(zdIs5) I; ivns-1(gk252) X | |||
| Genetic reagent (C. elegans) | CZ25508: Pmec-4-GFP(zdIs5) I; ivns-1(ok3171) X; Ex[ivns-1_gDNA(juEx7673)] | |||
| Genetic reagent (C. elegans) | CZ25509: Pmec-4-GFP(zdIs5) I; ivns-1(ok3171) X; Ex[ivns-1_gDNA(juEx7674)] | |||
| Genetic reagent (C. elegans) | CZ25510: Pmec-4-GFP(zdIs5) I; ivns-1(gk252) X; Ex[ivns-1_gDNA(juEx7673)] | |||
| Genetic reagent (C. elegans) | CZ25511: Pmec-4-GFP(zdIs5) I; ivns-1(gk252) X; Ex[ivns-1_gDNA(juEx7674)] | |||
| Genetic reagent (C. elegans) | CZ24755: juEx7584[Pesyt-2-GFP] | |||
| Genetic reagent (C. elegans) | CZ21465: Pmec-4-GFP(zdIs5) I; epi-1(gm121) IV | |||
| Genetic reagent (C. elegans) | CZ21463: Pmec-7-GFP(muIs32) II; emb-9(tk75) III | |||
| Genetic reagent (C. elegans) | CZ21198: Pmec-7-GFP(muIs32) II; adt-1(cn30) X | |||
| Genetic reagent (C. elegans) | CZ20937: Pmec-4-GFP(zdIs5) I; adt-2(wk156) X | |||
| Genetic reagent (C. elegans) | CZ21004: Pmec-4-GFP(zdIs5) I; adt-3 (T19D2.1) (ok923) X | |||
| Genetic reagent (C. elegans) | CZ26611: Pmec-7-GFP(muIs32) II; gon-1(e1254) IV / + | |||
| Genetic reagent (C. elegans) | CZ23908: rab-8(tm2526) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ23909: rab-11.2(tm2081) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ20682: Pmec-7-GFP(muIs32) II; nex-1(gk148) III | |||
| Genetic reagent (C. elegans) | CZ20683: Pmec-7-GFP(muIs32) II; nex-2(ok764) III | |||
| Genetic reagent (C. elegans) | CZ20684: Pmec-7-GFP(muIs32) II; nex-3(gk385) III | |||
| Genetic reagent (C. elegans) | CZ20685: Pmec-7-GFP(muIs32) II; nex-4(gk102) V | |||
| Genetic reagent (C. elegans) | CZ14006: Pmec-7-GFP(muIs32) II; mec-17(ok2109) IV | |||
| Genetic reagent (C. elegans) | CZ14008: Pmec-7-GFP(muIs32) II; atat-2(ok2415) X | |||
| Genetic reagent (C. elegans) | CZ14848: mec-17(ok2109) IV; atat-2(ok2415) X; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ17720: Pmec-7-GFP(muIs32) II; mec-12(tm5083) III | |||
| Genetic reagent (C. elegans) | CZ9247: tba-1(ok1135) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ26688: Pmec-4-GFP(zdIs5) I; tba-7(gk787939) III | |||
| Genetic reagent (C. elegans) | CZ26833: Pmec-4-GFP(zdIs5) I; tba-7(u1015) III | |||
| Genetic reagent (C. elegans) | CZ26635: Pmec-4-GFP(zdIs5) I; mec-7(ok2152) X | |||
| Genetic reagent (C. elegans) | CZ10615: Pmec-4-GFP(zdIs5) I; tbb-2(gk129) III | |||
| Genetic reagent (C. elegans) | CZ11083: Pmec-4-GFP(zdIs5) I; tbb-4(ok1461) X | |||
| Genetic reagent (C. elegans) | CZ10810: Pmec-4-GFP(zdIs5) I; tbb-6(tm2004) V | |||
| Genetic reagent (C. elegans) | CZ21461: Pmec-4-GFP(zdIs5) I; fli-1(ky535) III | |||
| Genetic reagent (C. elegans) | CZ21199: Pmec-7-GFP(muIs32) II; gsnl-1(ok2979) V | |||
| Genetic reagent (C. elegans) | CZ10888: viln-1(ok2413) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ13606: Pmec-4-GFP(zdIs5) I; twf-2(ok3564) X | |||
| Genetic reagent (C. elegans) | CZ20063: Pmec-4-GFP(zdIs5) I; mfsd-6(ju833) III | |||
| Genetic reagent (C. elegans) | CZ19827: Pmec-4-GFP(zdIs5) I; mfsd-6(tm3356) III | |||
| Genetic reagent (C. elegans) | CZ24417: Pmec-4-GFP(zdIs5) I; mfsd-6(tm3356) III; Prgef-1-mfsd-6(juEx6079) | |||
| Genetic reagent (C. elegans) | CZ21030: Pmec-4-GFP(zdIs5) I; C05D9.7(ok2931) X | |||
| Genetic reagent (C. elegans) | CZ25317: Pmec-4-GFP(zdIs5) I;dpy-10(e128) II | |||
| Genetic reagent (C. elegans) | CZ23667: Pmec-4-GFP(zdIs5) I;F35G2.1(ok1669) IV | |||
| Genetic reagent (C. elegans) | CZ23772: Pmec-4-GFP(zdIs5) I;gly-2(tm839) I | |||
| Genetic reagent (C. elegans) | CZ17890: Pmec-4-GFP(zdIs5) I;osm-11(rt142) X | |||
| Genetic reagent (C. elegans) | CZ17021: Pmec-4-GFP(zdIs5) I;zig-1(ok784) II | |||
| Genetic reagent (C. elegans) | CZ17023: Pmec-4-GFP(zdIs5) I;zig-3(gk33) X | |||
| Genetic reagent (C. elegans) | CZ17024: Pmec-4-GFP(zdIs5) I;zig-3(ok1476) X | |||
| Genetic reagent (C. elegans) | CZ22031: abts-1(ok1566) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ21461: Pmec-4-GFP(zdIs5) I; fli-1(ky535) III | |||
| Genetic reagent (C. elegans) | CZ17435: Pmec-7-GFP(muIs32) II;mec-12(e1605) III | |||
| Genetic reagent (C. elegans) | CZ17637: Pmec-4-GFP(zdIs5) I;tba-9(ok1858) X | |||
| Genetic reagent (C. elegans) | CZ20033: vab-10(e698) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ17099: Pmec-4-GFP(zdIs5) I; plk-1(or683ts) III | |||
| Genetic reagent (C. elegans) | CZ17285: Pmec-4-GFP(zdIs5) I; svh-2(tm737) X | |||
| Genetic reagent (C. elegans) | CZ19343: Pmec-4-GFP(zdIs5) I; cpr-1(ok1344) V | |||
| Genetic reagent (C. elegans) | CZ19200:dhhc-11(gk1105) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ22823: Pmec-4-GFP(zdIs5) I;eat-3(tm1107) II | |||
| Genetic reagent (C. elegans) | CZ16134: Pmec-4-GFP(zdIs5) I;npr-20(ok2575) II | |||
| Genetic reagent (C. elegans) | CZ23845: Pmec-4-GFP(zdIs5) I; ptps-1(tm1984) I | |||
| Genetic reagent (C. elegans) | CZ17995: supr-1(ju1118) I;Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ12031: Pmec-4-GFP(zdIs5) I;rsef-1(ok1356) X | |||
| Genetic reagent (C. elegans) | CZ17629: Pmec-4-GFP(zdIs5) I; osm-7(tm2256) III | |||
| Genetic reagent (C. elegans) | CZ17098: Pmec-4-GFP(zdIs5) I; par-2(or373ts) III | |||
| Genetic reagent (C. elegans) | CZ18676: Pmec-4-GFP(zdIs5) I; rgl-1(ok1921) X | |||
| Genetic reagent (C. elegans) | CZ20056: Pmec-7-GFP(muIs32) II; wdfy-3(ok912) IV | |||
| Genetic reagent (C. elegans) | CZ19721: Pmec-4-GFP(zdIs5) I wdr-23(tm1817) I | |||
| Genetic reagent (C. elegans) | CZ22063: Pmec-4-GFP(zdIs5) I; brap-2(tm5132) II | |||
| Genetic reagent (C. elegans) | CZ21217: Pmec-4-GFP(zdIs5) I; brap-2(ok1492) II | |||
| Genetic reagent (C. elegans) | CZ19337: Pmec-4-GFP(zdIs5) I; cdc-48.1(tm544) II | |||
| Genetic reagent (C. elegans) | CZ19725: Pmec-4-GFP(zdIs5) I; ced-9(n1950sd) III | |||
| Genetic reagent (C. elegans) | CZ21651: Pmec-4-GFP(zdIs5) I; dnj-23(tm7102) II | |||
| Genetic reagent (C. elegans) | CZ21356: Pmec-4-GFP(zdIs5) I; fbxc-50(tm5154) II | |||
| Genetic reagent (C. elegans) | CZ16950: Pmec-7-GFP(muIs32) II; math-33(ok2974) V | |||
| Genetic reagent (C. elegans) | CZ16951: Pmec-7-GFP(muIs32) II; skr-5(ok3068) V | |||
| Genetic reagent (C. elegans) | CZ21010: tep-1(tm3720) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ22796: mec-8(e398) I;Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ14510: Pmec-4-GFP(zdIs5) I;rict-1(mg360) II | |||
| Genetic reagent (C. elegans) | CZ22570: rtcb-1(gk451) I / [bli-4(e937) let-?(q782) qIs48](hT2) I, III; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ21655: Pmec-4-GFP(zdIs5) I; skn-1(ok2315) IV/nT1(qIs51) IV; V | |||
| Genetic reagent (C. elegans) | CZ23377: Pmec-4-GFP(zdIs5) I; smg-3(r930) IV | |||
| Genetic reagent (C. elegans) | CZ13997: Pmec-7-GFP(muIs32) II; syd-9(ju49) X | |||
| Genetic reagent (C. elegans) | CZ21723: Pmec-4-GFP(zdIs5) I; tdp-1(ok803) II | |||
| Genetic reagent (C. elegans) | CZ23133: Pmec-4-GFP(zdIs5) I; wdr-5.1(ok1417) III | |||
| Genetic reagent (C. elegans) | CZ21194: Pmec-7-GFP(muIs32) II; mig-17(k174) V | |||
| Genetic reagent (C. elegans) | CZ22792: Pmec-7-GFP(muIs32) II; ZC116.3(ok1618) V | |||
| Genetic reagent (C. elegans) | CZ19193: Pmec-7-GFP(muIs32) II;lgc-12(ok3546) III | |||
| Genetic reagent (C. elegans) | CZ18217: Pmec-4-GFP(zdIs5) I;tmc-1(ok1859) X | |||
| Genetic reagent (C. elegans) | CZ17639: Pmec-4-GFP(zdIs5) I;nud-1(ok552) III | |||
| Genetic reagent (C. elegans) | CZ17841: Pmec-4-GFP(zdIs5) I;mgl-1(tm1811) X | |||
| Genetic reagent (C. elegans) | CZ17843: Pmec-4-GFP(zdIs5) I;mgl-3(tm1766) IV | |||
| Genetic reagent (C. elegans) | CZ17848: Pmec-4-GFP(zdIs5) I;npr-25(ok2008) V | |||
| Genetic reagent (C. elegans) | CZ22890: Pmec-4-GFP(zdIs5) I;ucr-2.3(ok3073) III | |||
| Genetic reagent (C. elegans) | CZ15607: drag-1(tm3773) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ17393: Pmec-4-GFP(zdIs5) I; snb-6(tm5195) II | |||
| Genetic reagent (C. elegans) | CZ17018: Pmec-4-GFP(zdIs5) I; drag-1(tm3773) I | |||
| Genetic reagent (C. elegans) | CZ18617: Pmec-4-GFP(zdIs5) I; ect-2(ku427) II | |||
| Genetic reagent (C. elegans) | CZ18818: Pmec-4-GFP(zdIs5) I; lin-2(e1309) X | |||
| Genetic reagent (C. elegans) | CZ18817: Pmec-4-GFP(zdIs5) I; magi-1(zh66) IV | |||
| Genetic reagent (C. elegans) | CZ20673: Pmec-4-GFP(zdIs5) I; prmt-5(gk357) III | |||
| Genetic reagent (C. elegans) | CZ18816: Pmec-4-GFP(zdIs5) I; rap-1(tm861) IV | |||
| Genetic reagent (C. elegans) | CZ18460: Pmec-4-GFP(zdIs5) I; smz-1(ok3576) IV | |||
| Genetic reagent (C. elegans) | CZ22544: Pmec-4-GFP(zdIs5) I; trxr-1(sv47) IV | |||
| Genetic reagent (C. elegans) | CZ24963: Pmec-4-GFP(zdIs5) I; natb-1(ju1405) V/nT1 IV; V | |||
| Genetic reagent (C. elegans) | CZ16946: Pmec-7-GFP(muIs32) II; rnf-5(tm794) III | |||
| Genetic reagent (C. elegans) | CZ23068: ulp-5/tofu-3(tm3063) I; Pmec-7-GFP(muIs32) II | |||
| Genetic reagent (C. elegans) | CZ23091: Pmec-7-GFP(muIs32) II; csr-1(fj54) IV/nT1 IV; V | |||
| Genetic reagent (C. elegans) | CZ17638: Pmec-4-GFP(zdIs5) I; elpc-3(ok2452) V | |||
| Genetic reagent (C. elegans) | CZ12938: Pmec-4-GFP(zdIs5) I; hda-6(tm3436) IV | |||
| Recombinant DNA reagent | Plasmid: pCZGY3260:nmat-2 genomic DNA | This work | N/A | nmat-2 genomic DNA (~1500 bp upstream;~670 bp downstream); modified pCFJ201 plasmid for modified MosSCI on ChIV |
| Recombinant DNA reagent | Plasmid: pCZ993:Pmec-4-nmat-2 gDNA-let-858 3'UTR | This work | N/A | nmat-2 expression driven by mec-4 promoter in the mechanosensory neurons |
| Recombinant DNA reagent | Plasmid: pCZ994: Pmtl-2-nmat-2 gDNA-let-858 3'UTR | This work | N/A | nmat-2 expression driven by mtl-2 promoter in the intestine |
| Recombinant DNA reagent | Plasmid: pCZ995: Pcol-12-nmat-2 gDNA-let-858 3'UTR | This work | N/A | nmat-2 expression driven by col-12 promoter in the epiderdims |
| Recombinant DNA reagent | Plasmid: pCZGY3329: Pmec-4-GFP-ESYT-2 | This work | N/A | |
| Recombinant DNA reagent | Plasmid: pCZGY3344: Pmec-4-mKate2-ESYT-2 | This work | N/A | |
| Recombinant DNA reagent | Plasmid: pCZGY3342: Pmec-4-mKate2-PISY-1 | This work | N/A | |
| Recombinant DNA reagent | Plasmid: pCZGY3302: ivns-1 genomic DNA | This work | N/A | ivns-1 genomic DNA (2 kb upstream; 800 bp downstream) |
| Recombinant DNA reagent | Plasmid: pCZGY3347: Prgef-1-mfsd-6 | This work | N/A | |
| Recombinant DNA reagent | Plasmid: pCZGY3346:Pesyt-2-GFP | This work | N/A | GFP expression driven by esyt-2 promoter |
| Sequence- based reagent | crRNA: nmat-2:/AltR1/rCrGrArGrU rCrGrC rUrCrU rUrCrU rUrGrC rCrGrUrGrUrU rUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to makenmat-2(ju1512) and nmat-2(ju1514) |
| Sequence- based reagent | crRNA: nmat-2:/AltR1/rCrGrUrGrU rUrGrA rArCrU rArArC rUrCrC rArCrU rGrUrU rUrUrA rGrArG rCrUrArUrGrC rU/AltR2/ | IDT | N/A | crRNA to makenmat-2(ju1512) |
| Sequence- based reagent | crRNA: nmat-1:/AltR1/rArA rCrUrU rUrUrU rCrGrGrUrCrC rCrCrA rUrArG rGrUrU rUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to mak e nmat-1(ju1565) |
| Sequence- based reagent | crRNA: nmat-1:/AltR1/rArU rGrUrA rCrUrU rGrArU rUrArC rGrGrA rArUrC rGrUrU rUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to make nmat-1(ju1565) |
| Sequence- based reagent | crRNA: qns-1:/AltR1/rGrGrUrGrU rUrArU rUrCrA rCrGrU rGrUrU rArCrA rGrUrU rUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to make qns-1(ju1563) |
| Sequence-based reagent | crRNA: qns-1:/AltR1/rGrA rUrArA rCrUrG rArArA rUrCrU rGrGrA rUrArG rGrUrU rUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to make qns-1(ju1563) |
| Sequence-based reagent | crRNA: esyt-2:/AltR1/rGrG rUrUrU rCrArG rUrArA rUrUrGrUrGrG rGrCrU rGrUrUrUrUrA rGrArG rCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to make esyt-2(ju1409) |
| Sequence-based reagent | crRNA: esyt-2:/AltR1/rGrU rGrCrA rCrUrU rArCrG rGrGrU rUrGrU rArGrGrGrUrU rUrUrA rGrArGrCrUrA rUrGrC rU/AltR2/ | IDT | N/A | crRNA to makeesyt-2(ju1409) |
| Peptide, recombinant protein | Protein: Cas9-NLS purified protein | QB3 MacroLab, UC Berkley | N/A | |
| Peptide, recombinant protein | Phusion High-Fidelity DNA polymerases | Thermo Scientific | Cat#F530L | |
| Peptide, recombinant protein | DreamTaq DNA polymerases | Thermo Scientific | Cat#EP0705 | |
| Commercial assay or kit | ||||
| Chemical compound, drug | 5-fluoro-2-deoxy uridine | Sigma-Aldrich | Cat#50-91-9 | |
| Software, algorithm | ImageJ | NIH image | RRID:SCR_003070 | |
| Software, algorithm | ZEN | Zeiss | https://www.zeiss.com/microscopy/us/downloads/zen.html | |
| Software, algorithm | Zeiss LSM Data Server | Zeiss | https://www.zeiss.com/microscopy/us/downloads/lsm-5-series.html | |
| Software, algorithm | GraphPad Prism 5 | GraphPad Software, Inc. | RRID:SCR_002798 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39756.019





