The allosteric activation of cGAS underpins its dynamic signaling landscape
Figures
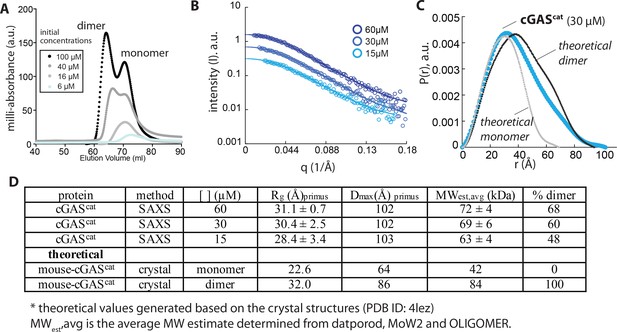
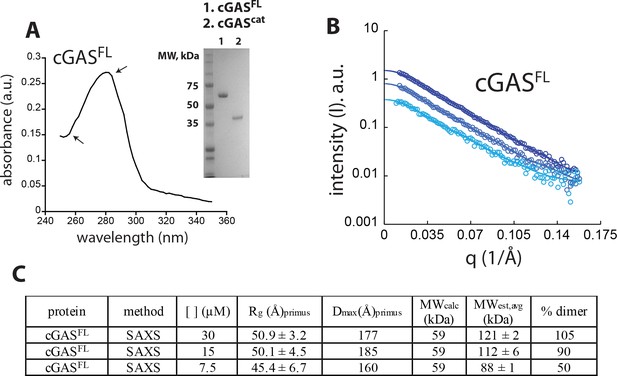
Human wild-type cGAScat can dimerize on its own.
(A) SEC (Superdex 75 16/600) profile of cGAScat. (B) SAXS scattering profile of cGAScat. (C) Pair-wise distance distribution functions of cGAScat. Theoretical P(r)s from mouse-cGAScat are shown for comparison (PDB ID: 4lez). (D) Summary of SAXS experiments.
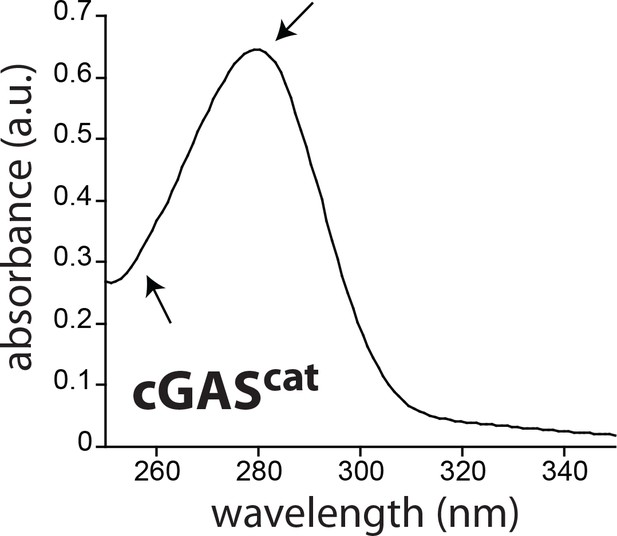
Uv-vis absorbance profile of purified cGAScat.
The predominant peak at 280 nm and the lack thereof at 260 nm consistently indicate no significant nucleic acid contamination.
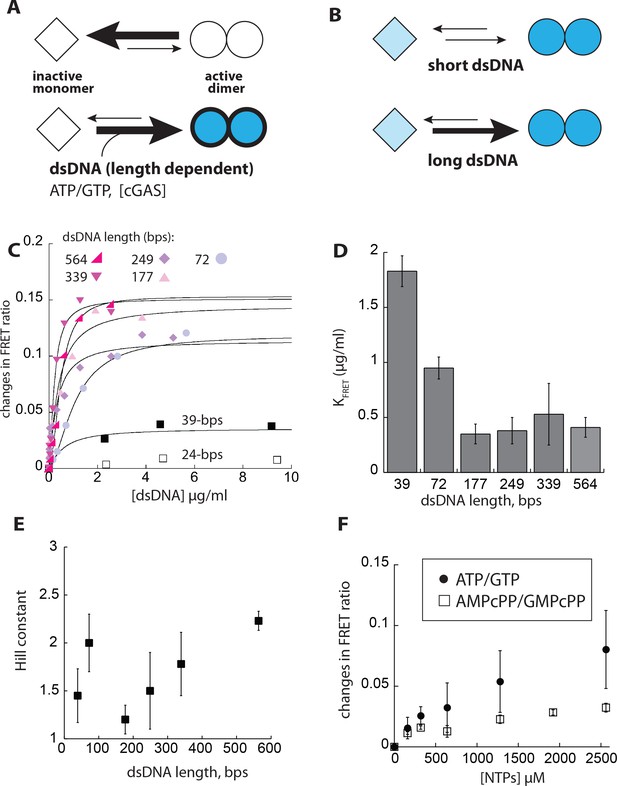
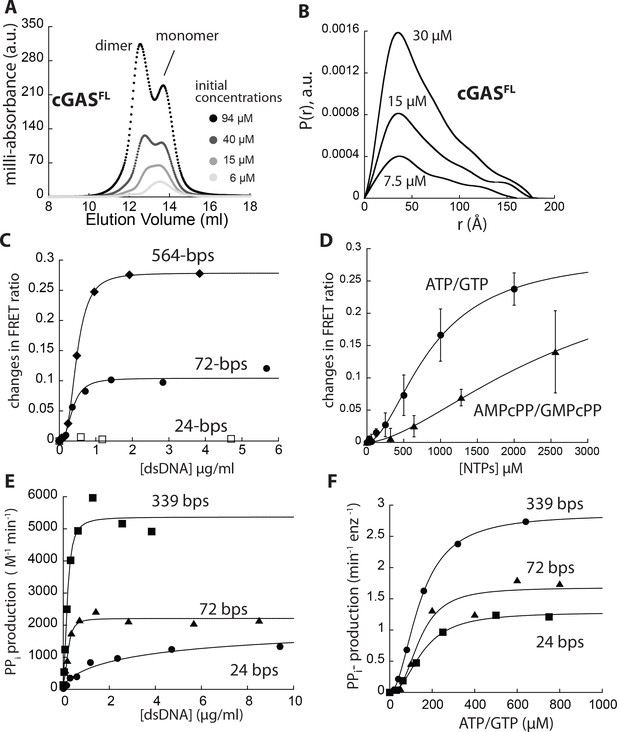
dsDNA cooperatively induces the dimerization of cGAS in a length-dependent manner.
(A) A scheme describing the allosteric framework of cGAS activation. Here, cGAS is subject to an intrinsic allosteric equilibrium with two major activity/conformational states, namely inactive monomer and active dimer. Resting cGAS is predominantly an inactive monomer (top). dsDNA (length-dependent) binding, increasing cGAS concentration, and substrate binding synergistically drive the allosteric equilibrium toward the active dimer. (B) An allosteric model describing dsDNA length-dependent distribution of active dimers and inactive (basally active) monomers. (C) Changes in the ratio between FRET donor emission (λmax: 578 nm) and the acceptor emission (λmax: 678 nm) of labeled cGAScat (20 nM each) at indicated dsDNA concentrations. (D) A plot of dimerization efficiency (KFRET) vs. dsDNA length (n = 3;±SD). (E) A plot of fitted Hill constants vs. dsDNA lengths (n = 3;±SD). (F) Changes in the ratio between the FRET donor/acceptor emission ratios of labeled cGAScat (20 nM each) at indicated NTP pair concentrations. (n = 3;±SD).
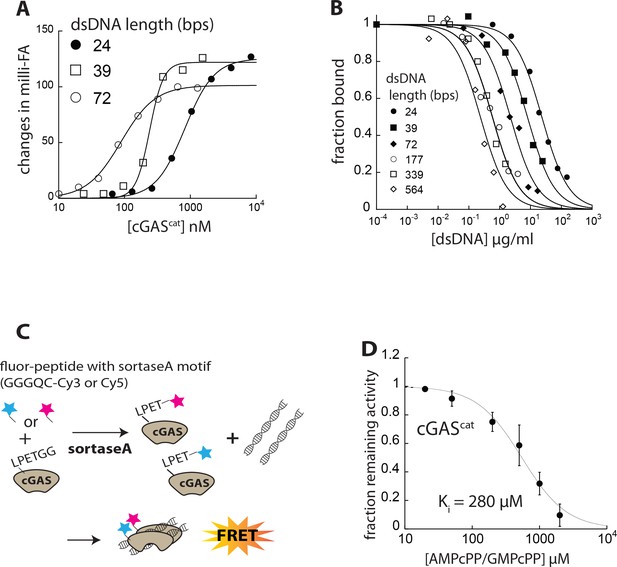
dsDNA binding and dimerization of cGAS.
(A) Binding of cGAScat to each FAM-labeled dsDNA (5 nM) was determined by FA. The lines are fits to a standard binding isotherm. (B) Competition binding assay using 72 bp FAM-dsDNA (5 nM) and cGAScat (80 nM;~KD for 72 bp FAM-dsDNA) against various unlabeled dsDNA lengths; the lines are fits to a competition binding equation: (1/ (1+(dsDNAcompetitor)/IC50)Hill constant). Fraction bound was calculated based on the changes in fluorescence anisotropy (FA) of 72 bp FAM-dsDNA. (C) Scheme for FRET-based dimerization assay. (D) Competition activity assay using 339 bp dsDNA (4 µg/ml) and cGAScat (50 nM) and 150 µM ATP/GTP against increasing concentrations of AMPcPP/GMPcPP; the lines are fits to a competition binding equation: (1/ (1+(competitor)/IC50)Hill constant). The Reported Ki was calculated using the Cheng-Prusoff equation (Ki = IC50/([1+([S]/KM)]).
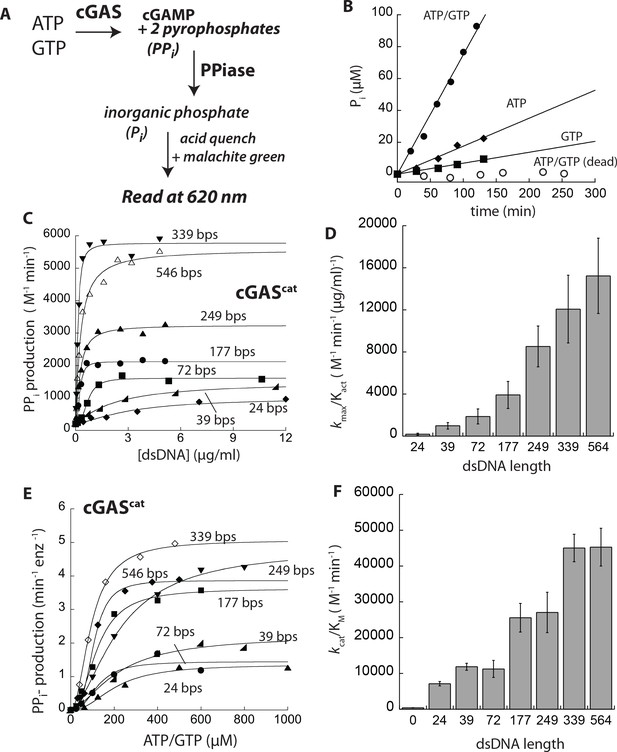
dsDNA length can grade the activity of cGAS.
(A) PPiase-coupled assay scheme. (B) A plot of time-dependent Pi production of cGAScat (125 nM) at various conditions (dead: E225A-D227A). (C) A plot of the dsDNA concentration-dependent NTase activity of cGAScat (25 nM) and 1 mM ATP/GTP with various duplex lengths. Lines are fits to a Hill form of the Michaelis-Menten equation. (D) A plot of dsDNA-affinity normalized maximal activities of cGAScat vs. dsDNA lengths (n = 3;±SD). (E) Substrate dependence of the steady-state rate of NTase activity by cGAScat (125 nM) with saturating amounts of each dsDNA (6X Kact). Lines are fits to a Hill form of the Michaelis-Menten equation. (F) A plot of catalytic efficiencies (kcat/KM) vs. dsDNA lengths (n = 3;±SD).

Kinetic parameters of cGAS and PPiase.
(A) Steady-state kinetics of PPiase. Calculated kcat/KM is ~675,000 M−1sec−1, which is about 1,000-fold faster than that of cGAS in the presence of saturating 564 bp dsDNA (~750 M−1sec−1). (B) Steady-state NTase rates of dsDNA free cGAScat (estimated kcat/KM ≈ 800 M−1min−1).
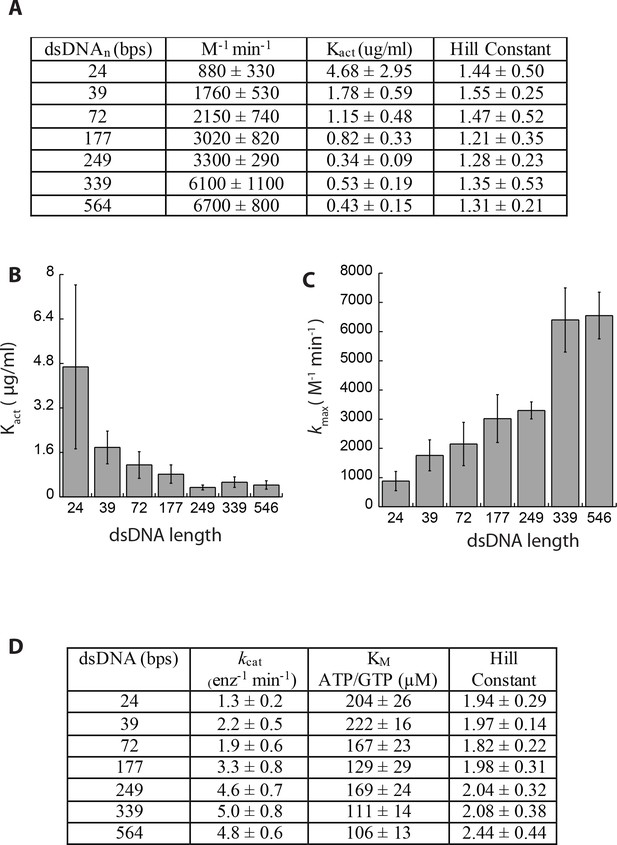
Summary of activation parameters.
(A) Table summarizing activation parameters. (B) A plot of the half-maximal dsDNA concentrations (Kact) required to activate cGAS (i.e. apparent binding affinity) vs. dsDNA lengths. (C) A plot of the maximal dsDNA induced NTase activity of cGAS (kmax) vs. dsDNA lengths. (D) Table summarizing steady-state kinetic parameters.
cGASFL operates within the same allosteric framework as cGAScat.
(A) SEC of cGASFL (Superdex 200 10/300). (B) Pair-wise distance distribution function of cGASFL. (C) Changes in the ratio between the FRET donor emission (λmax: 578 nm) and the acceptor emission (λmax: 678 nm) of labeled cGASFL (20 nM each) at indicated dsDNA concentrations. (D) Changes in the ratio between the FRET donor/acceptor emission ratio of labeled cGASFL (20 nM each) at indicated NTP pair concentrations. (E) A plot of the dsDNA concentration-dependent NTase activity of cGASFL (25 nM) and 800 µM ATP/GTP with various duplex lengths. Lines are fits to a Hill form of the Michaelis-Menten equation. (F) Substrate dependence of the steady-state rate of NTase activity by cGASFL (125 nM) with saturating amounts of each dsDNA (6X Kact). Lines are fits to a Hill form of the Michaelis-Menten equation.
Biophysical properties of cGASFL.
(A) Uv-vis spectra of cGASFL showing no significant nucleic acid contamination, and SDS-PAGE of cGAScat and cGASFL. SDS-PAGE gel shows >95% pure cGASFL and cGAScat. (B) SAXS scattering profile of cGASFL. (C) Table summarizing SAXS analyses of cGASFL.
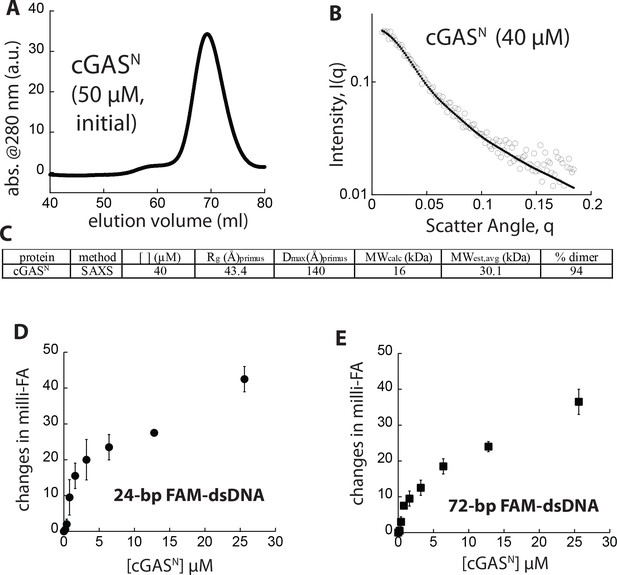
Biophysical properties of cGASN.
(A) SEC of cGASN (Superdex 75 16/600). (B) SAXS scattering profile of cGASN. (C) Table summarizing SAXS analyses of cGASFL. (D-E) Binding of cGASN to each FAM-labeled dsDNA (5 nM) was monitored via FA.
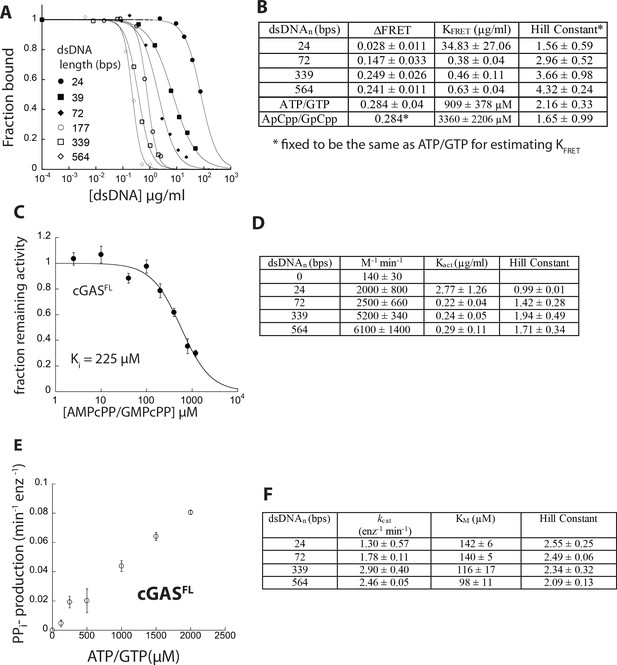
dsDNA binding and dimerization activities of cGASFL.
(A) Competition binding assay using 72 bp FAM-dsDNA (5 nM) and cGASFL (50 nM) against various dsDNA lengths; the lines are fits to a competition binding equation: (1/ (1+(dsDNAcompetitor)/IC50)Hill constant). (B) Table summarizing the FRET dimerization assay results of cGASFL. (C) Competition activity assay using 339 bp dsDNA (4 µg/ml) and cGAScat (50 nM) and 150 µM ATP/GTP against increasing concentrations of AMPcPP/GMPcPP; the lines are fits to a competition binding equation: (1/ (1+(competitor)/IC50)Hill constant. The Ki was calculated using the Cheng-Prusoff equation. (D) Table summarizing activation parameters of cGASFL (25 nM, 800 µM ATP/GTP). (E) Steady-state NTase rates of dsDNA free cGASFL (estimated kcat/KM ≈ 400 M−1 min−1). (F) Table summarizing steady-state kinetic parameters of cGASFL (125 nM, [dsDNA]=6 x Kact).
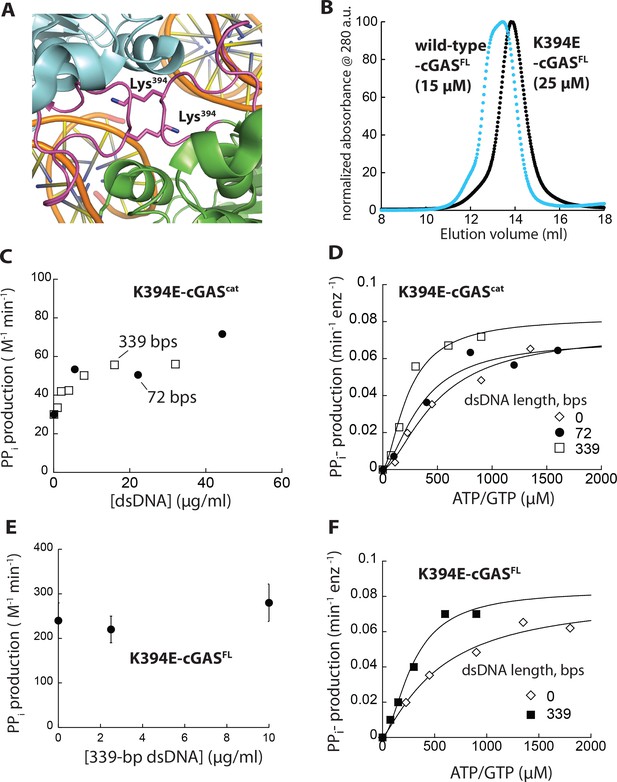
Monomeric cGAS is basally active, but cannot be further activated by dsDNA.
(A) Crystal structure of dimeric cGAScat. The loop important for dimerization is colored in magenta and Lys394 is shown in stick representation (PDB ID: 4lez). (B) SEC (Superdex 200 10/300) of K394E-cGASFL. WT-cGASFL is shown for reference (blue). (C) A plot of the dsDNA-concentration dependent NTase activity of K394E-hcGAScat (1 µM) and 1 mM ATP/GTP with different duplex lengths. (D) Substrate dependence of the steady-state rate of NTase activity by K394E-hcGAScat (1 µM) in the absence or presence of each dsDNA (6X Kact). Lines are fits to a Hill form of Michaelis-Menten equation. (E) A plot of the dsDNA-concentration dependent NTase activity of K394E-cGASFL (125 nM) and 1 mM ATP/GTP with different duplex lengths. (F) Substrate dependence of the steady-state rate of NTase activity by K394E-cGASFL (125 nM) in the absence or presence of each dsDNA (6X Kact). Lines are fits to a Hill form of Michaelis-Menten equation.
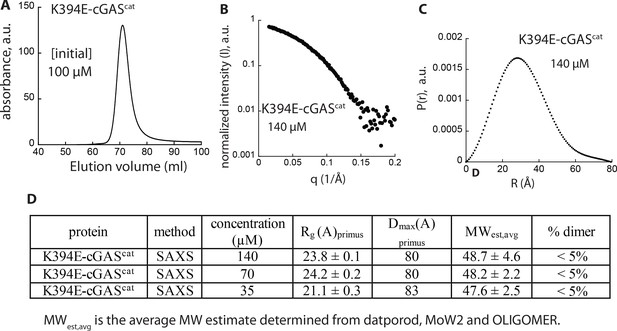
Biophysical properties of K394E-cGAS.
(A) SEC of K394E-cGAScat (Superdex75 16/600). (B) SAXS scattering profile of K394E-cGAScat. (C) Pair-wise distance distribution function calculated from Figure 5—figure supplement 1B. (D) Table summarizing SAXS analyses of K394E-cGAScat.
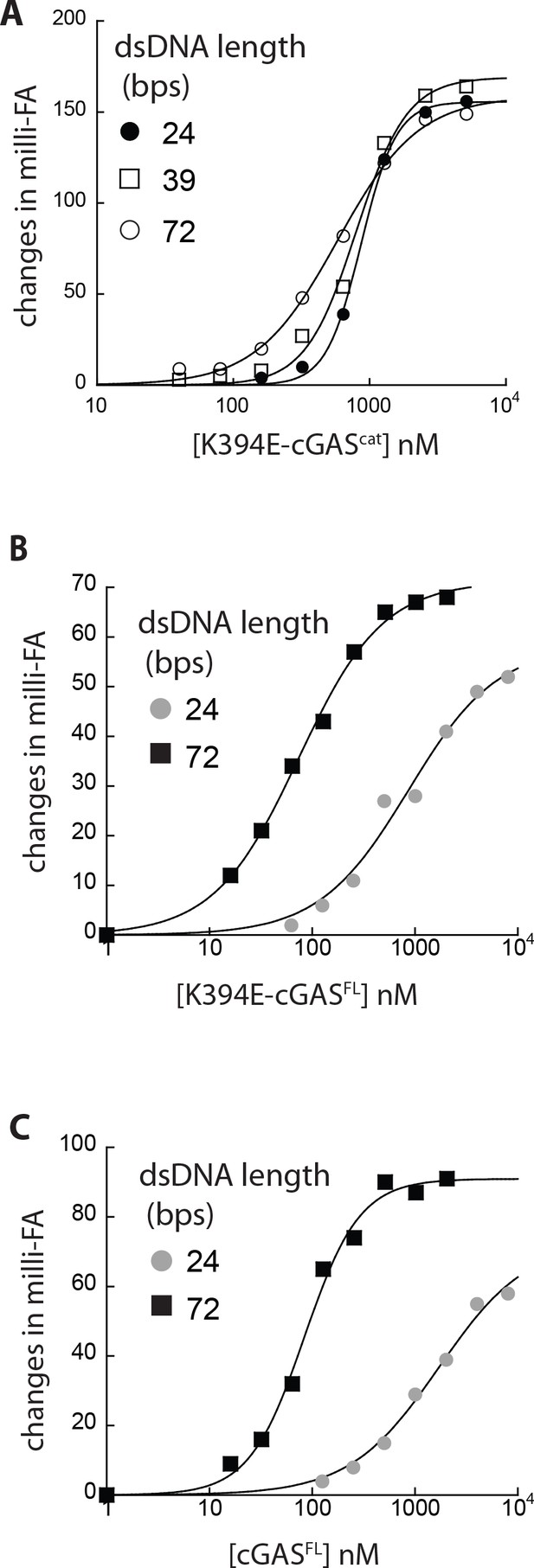
dsDNA binding properties of K394-cGAS.
(A) Binding of K394E-cGAScat to each FAM-labeled dsDNA (5 nM) was determined by FA. The lines are fits to a standard binding isotherm. (B) Binding of K394E-cGASFL to each FAM-labeled dsDNA (5 nM) was determined by fluorescence anisotropy (FA). The lines are fits to a standard binding isotherm. (C) Binding of cGASFL to each FAM-labeled dsDNA (5 nM) was determined by fluorescence anisotropy (FA). The lines are fits to a standard binding isotherm.
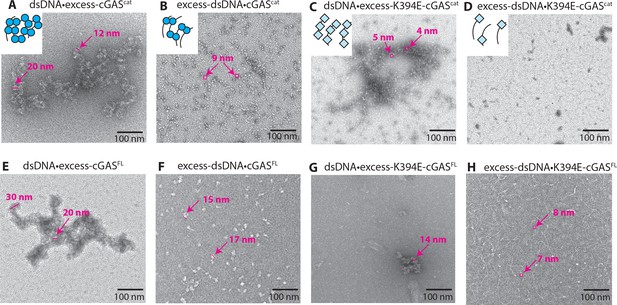
cGAS dimers assume various configurations on dsDNA.
Negative-stain electron micrographs of cGAS•dsDNA complexes. (A, E) 3-fold excess cGAS over dsDNA, (B, F) 3-fold excess dsDNA over cGAS, (C, G) 3-fold excess K349E-cGAS over dsDNA, (D, H) 3-fold excess dsDNA over K394E-cGAS. Ratios of protein to dsDNA or dsDNA to protein are binding site normalized; 18 bp per binding site. The particle sizes in B and F are consistent with the Dmax of cGAScat and cGASFL, respectively (Figures 1 and 4). Particle sizes in C and H are consistent with the Dmax of K394E-cGAS variants (Figure 5—figure supplement 1).

Additional nsEM images.
(A-E) Zoomed-in images from Figure 6B,C,F and H showing the measured particles.
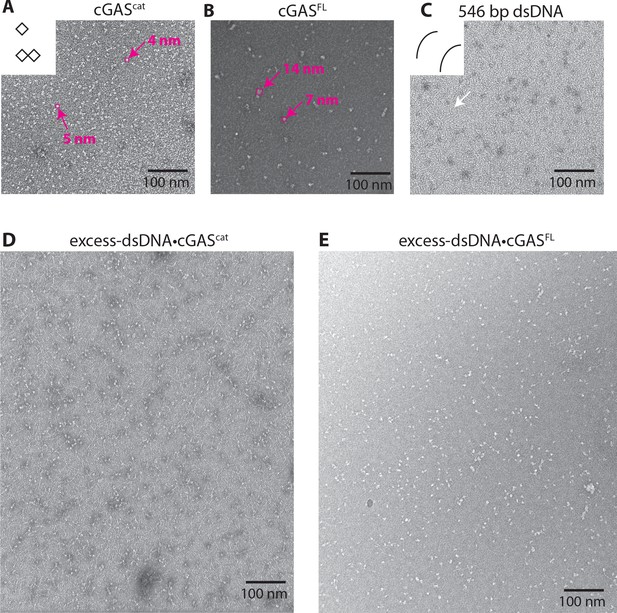
Additional nsEM images.
(A-E) Additional negative-stain electron micrograph images.
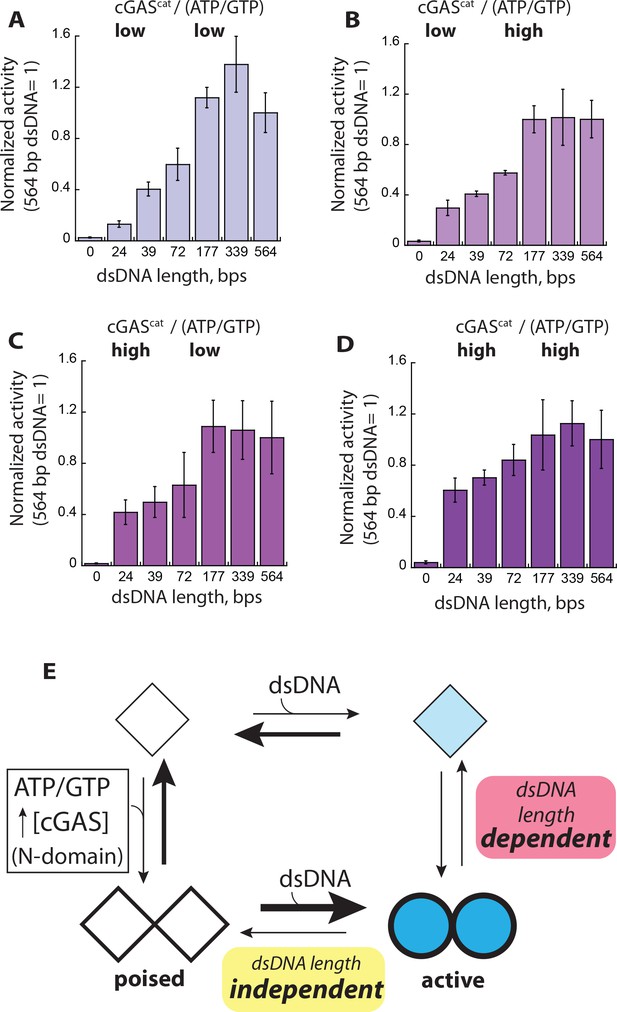
Context dependent activation of cGAS.
(A–D) Plots of the NTase activity of cGAScat vs. dsDNA length (6X-Kact) at various enzyme and substrate concentrations (low cGAS: 50 nM, High cGAS: 1 µM; low ATP/GTP: 250 µM, high ATP/GTP: 6X-KM for each dsDNA). The NTase activity of cGAScat induced by 564 bp dsDNA was normalized by enzyme and substrate concentrations, and used as the reference to calculate the fraction activity for each dsDNA length (n = 3; ±SD). (E) The equilibrium-based activation model of cGAS. Diamonds represent basally active cGAS and circles indicate active cGAS. Filled shapes and thicker lines indicate dsDNA binding and substrate binding, respectively. Thicker lines and darker shades indicate stronger interactions.
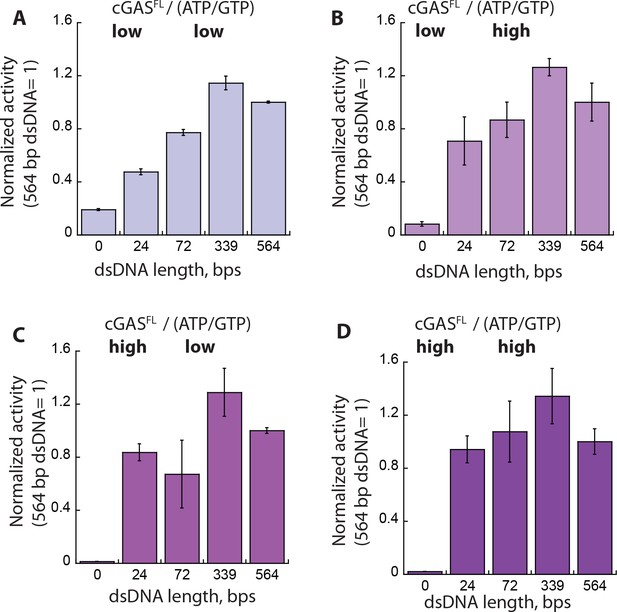
Context dependent activation of cGASFL.
(A-D) Plots of the NTase activity of cGASFL vs. dsDNA length (6X-Kact) at various enzyme and substrate concentrations (low cGAS: 25 nM, High cGAS: 1 µM; low ATP/GTP: 150 µM, high ATP/GTP: 6X-KM for each dsDNA). The NTase activity of cGASFL induced by 564 bp dsDNA was normalized by enzyme and substrate concentrations, and used as the reference to calculate the fraction activity for each dsDNA length (n = 3;±SD).
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39984.022





