Meiotic drive of female-inherited supernumerary chromosomes in a pathogenic fungus
Figures
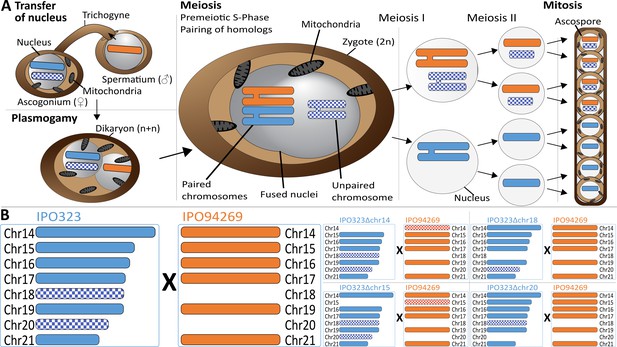
Meiosis and chromosome segregation in Z.tritici.
(A) Schematic overview of the assumed sexual process between two parental strains of Z. tritici (Alexopoulos et al., 1996; Kema et al., 2018) with one supernumerary chromosome shared and therefore paired (blue/orange) and one supernumerary chromosome unique to one strain (blue checkered) and unpaired in the zygote. The spermatial nucleus is transferred from the male partner via the trichogyne to the ascogonium of the female partner, resulting in plasmogamy and a dikaryon with two separate nuclei. Prior to karyogamy, the chromosomes are replicated and thus comprise each two chromatids when meiosis is initiated by pairing of homologous chromosomes. In meiosis I, homologous chromosomes are segregated, followed by chromatid separation in meiosis II. A subsequent mitosis results in the production of eight ascospores contained within one ascus. The expected segregation of chromosomes according to Mendelian law of segregation is shown - which for unpaired chromosomes is 4:0. (B) Schematic illustration of the distribution of supernumerary chromosomes present in the parental strains exemplified for five of nine different crosses performed in this study. Parental strain IPO323 contains eight supernumerary chromosomes (chr14-21, blue). Parental strain IPO94269 contains six supernumerary chromosomes with homologs in IPO323 (chr14, chr15, chr16, chr17, chr19, and chr21 in orange). The IPO323 chromosomes chr18 and chr20 are not present in IPO94269. We used a set of IPO323 chromosome deletion strains to generate an additional unpaired chromosome (as examples IPO323∆chr14 X IPO94269, IPO323∆chr15 X IPO94269, IPO323∆chr18 X IPO94269, IPO323∆chr20 X IPO94269 to demonstrate the one to three unpaired chromosomes and the five to six paired chromosomes present in the different crosses). Orange and blue indicate chromosomes that are shared between both strains. Checkered orange and checkered blue indicate chromosomes that are unique to one parent and therefore unpaired in the zygote.
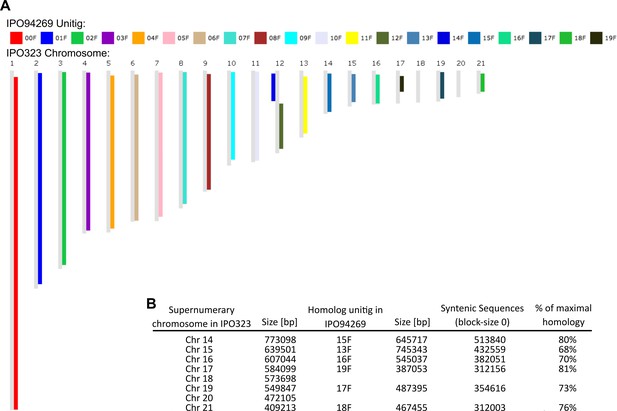
Synteny comparison of IPO94269 and IPO323.
(A) Synteny blot for IPO94269 unitigs on IPO323 chromosomes. (B) Summary table of syntenic regions of IPO323 and IPO94269.
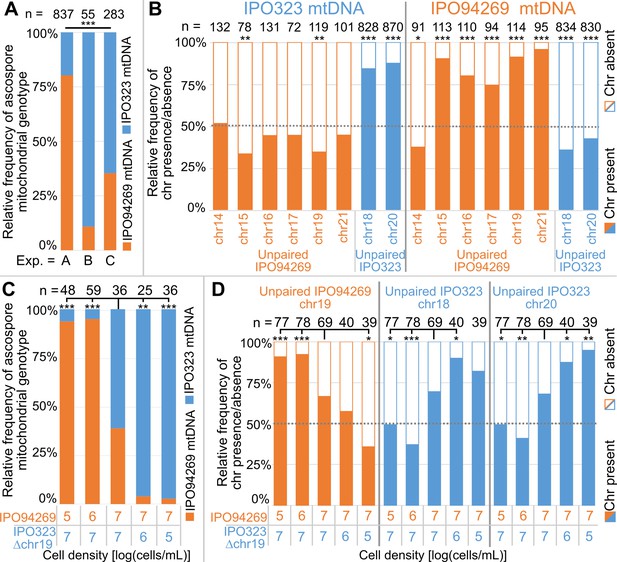
Unpaired supernumerary chromosomes show a segregation advantage only when inherited from the female parent.
(A) Relative frequencies of mitochondrial genotypes in random and randomized ascospores in experiments A, B and C. The mitochondrial transmission varied significantly between the three experiments. Statistical significance was inferred by Fisher’s exact test (p<2.2*10−16). (B) Relative frequencies of the presence and absence of unpaired supernumerary chromosomes in all progeny ascospores pooled for experiments A, B and C according to the mitochondrial genotype of the ascospore. Orange and blue indicate unpaired chromosomes originating from IPO94269 or IPO323, respectively. Unpaired chromosomes (with the exception of chr14) inherited from the parent that provided the mitochondrial genotype (i.e. the female parent) are overrepresented in the progeny, while the same chromosomes when originating from the male parent are not. Statistical significance was inferred by a two-sided binomial test with a probability of p=0.5. (C) Cell density affects the sexual role during mating and thereby the transmission of the mitochondria. Relative frequencies of mitochondrial genotype in random and randomized ascospores isolated from crosses of IPO94269 and IPO323Δchr19 that were co-inoculated on wheat at different cell densities. The resulting progeny shows a correlation between cell-density and mitochondria transmission. Strains inoculated at lower density in general take the female role as observed by the mitochondrial transmission. Statistical significance was inferred by a two-sided Fisher’s exact test compared to the co-inoculation with equal cell densities of both strains. (D) The cell density affects the transmission of unpaired chromosomes. Relative frequencies in all ascospores of the presence and absence of unpaired supernumerary chromosomes 19, inherited from parent IPO94269 and unpaired chromosomes 18 and 20, inherited from IPO323 Δchr19 are indicated according to the cell density of the parental strains IPO94269 and IPO323Δchr19 at inoculation. Statistical significance was inferred by a two-sided Fisher’s exact test compared to the co-inoculation with equal cell density of both strains. (*=p < 0.05, **=p < 0.005, ***=p < 0.0005, see Supplementary file 5 for details on all statistical tests).
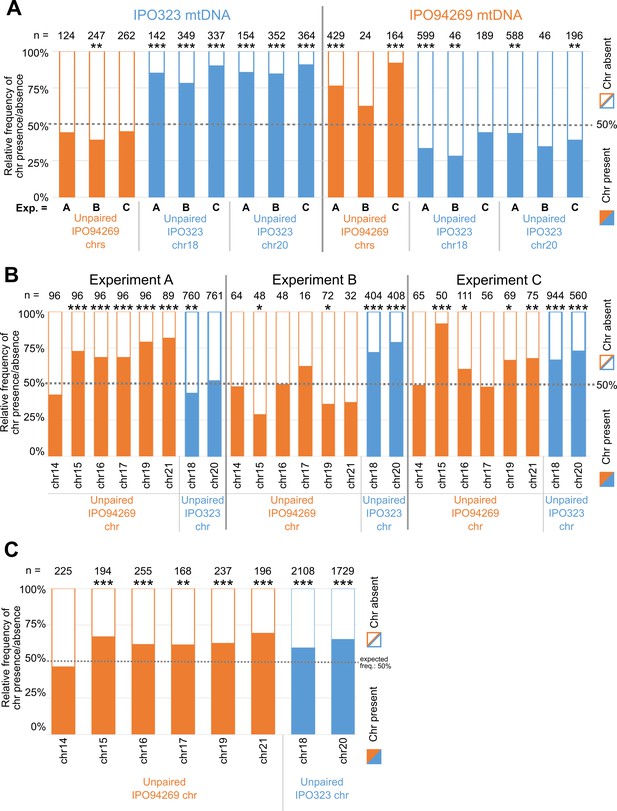
Transmission of unpaired chromosomes and mitochondria.
(A) Detailed presence/absence frequencies for all unpaired supernumerary chromosomes according to the mitochondrial genotype present in all ascospores. Data for the three experiments A, B and C are depicted separately. Statistical significance was inferred by a two-sided binomial test with a probability of p=0.5. (B) Detailed presence/absence frequencies for all unpaired supernumerary chromosome in experiment A, B and C, restricted to all co-inoculation crosses using 1*107 cells/mL cell density for both parental strains. Experiment A, B and C differ in the observed transmission advantage for the chromosomes originating from the parent IPO94269 (orange) or IPO323 (blue). Statistical significance was inferred by a two-sided binomial test with a probability of p=0.5. (C) Presence/absence frequencies of all unpaired supernumerary chromosomes pooled for all three experiments A, B and C, restricted to all co-inoculation crosses using the 1*107 cells/mL cell density for both parental strains. All supernumerary chromosomes, with the exception of chromosome 14 show a highly significant transmission advantage. Statistical significance was inferred by a two-sided binomial test with a probability of p=0.5. (*=p < 0.05, **=p < 0.005, ***=p < 0.0005, see Supplementary file 5 for details on all statistical tests).
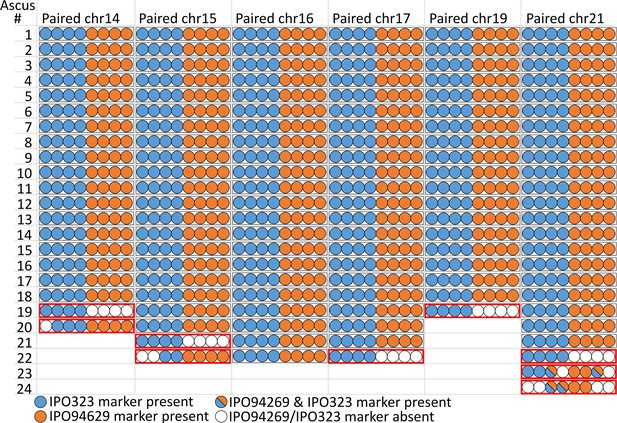
Paired supernumerary chromosomes show Mendelian segregation with frequent losses.
Analysis of segregation of paired supernumerary chromosomes in 24 complete tetrads. The transmission of chromosomes 14, 15, 16, 17, 19, and 21 with homologs in both parental strains IPO94269 and IPO323 was detected using segregating markers for chromosomes inherited from the parental strains IPO94269 (orange) and IPO323/IPO323∆chr14-21 (blue) in the eight ascospores originating from 24 asci. In 120 of the 129 cases (black outline) we observed a 4:4 ratio in the progeny. Note: for crosses/chromosome combinations where no paired chromosome was present no ascus is shown, which reduces the number of shown asci from the 24 asci that were analyzed in both Figure 3 and Figure 4.
-
Figure 3—source data 1
Examples of gel electrophoresis of PCR products for (A) core chromosome 13 marker on mating type, (B) core chromosome 4 marker 11O21, (C) core chromosome 4 marker 04L20, (D) core chromosome three marker caa-0002, core chromosome five marker ggc-001, core chromosome seven marker ac-001, (E) segregating marker of the mitochondrial genotype, (F) segregating marker of the supernumerary chromosome 14, (G) segregating marker of the supernumerary chromosome 15, (H) segregating marker of the supernumerary chromosome 16, (I) segregating marker of the supernumerary chromosome 17, (J) segregating marker of the supernumerary chromosome 19, (K) segregating marker of the supernumerary chromosome 19, (L) subtelomeric marker of the supernumerary chromosome 19, (M) subtelomeric marker of the supernumerary chromosome 19, (N) segregating marker of the supernumerary chromosome 21, (O) subtelomeric marker of the supernumerary chromosome 18, (P) centromeric marker of the supernumerary chromosome 18, (Q) subtelomeric marker of the supernumerary chromosome 18, (R) subtelomeric marker of the supernumerary chromosome 20, (S) centromeric marker of the supernumerary chromosome 20, (T) subtelomeric marker of the supernumerary chromosome 20.
- https://doi.org/10.7554/eLife.40251.009
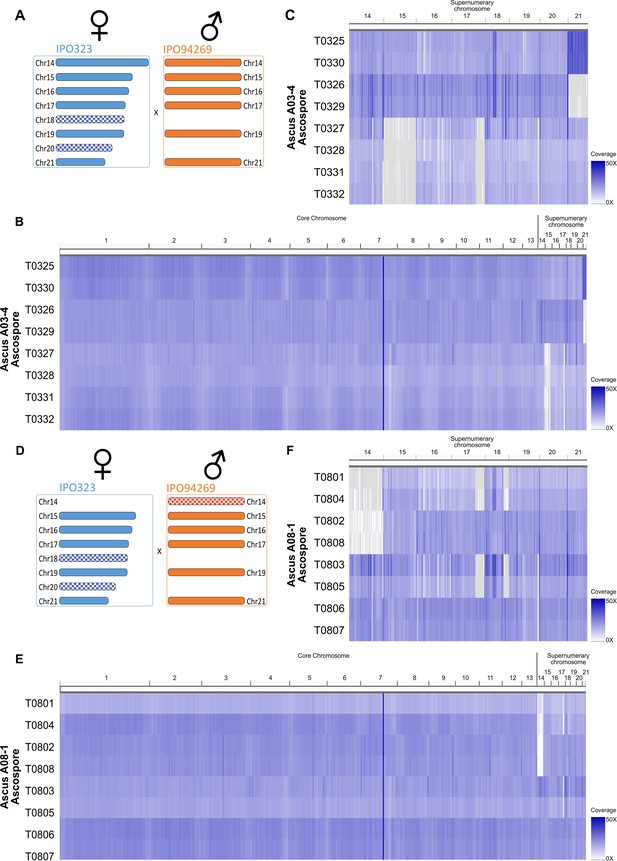
Whole genome sequencing confirms chromosome drive for unpaired supernumerary chromosomes.
Coverage heatmap for eight ascospores (T0325-T0332) of ascus A03-4 (A–C) and eight ascospores (T0801-T0808) of ascus A08-1 (D–F). (a) Supernumerary chromosome complement in cross resulting in ascus A03-4 between IPO323 and IPO94269. (B) Coverage heatmap of all chromosomes on from ascus A03-4 reflecting similar coverage for essential and supernumerary chromosomes. (C) Coverage heatmap of supernumerary chromosomes of ascus A03-4 indicating constant coverage of chromosome 18 and 20 in all eight ascospores, loss of paired chromosome 15 in four ascospores and non-disjunction of sister-chromatids in meiosis two resulting in two ascospores containing two chromosomes 21 and two corresponding ascospores lacking chromosome 21. (D) Supernumerary chromosome complement in cross resulting in ascus A08-1 between PO323∆chr14 and IPO94269. (E) Coverage heatmap of all chromosomes on from ascus A08-1 reflecting similar coverage for essential and supernumerary chromosomes. (F) Coverage heatmap of supernumerary chromosomes of ascus A08-1 indicating constant coverage of chromosome 20 in all eight ascospores, segregating unpaired chromosome 14 inherited from the male parental strain and segregating loss of coverage on right arm of chromosome 18 in four ascospores.
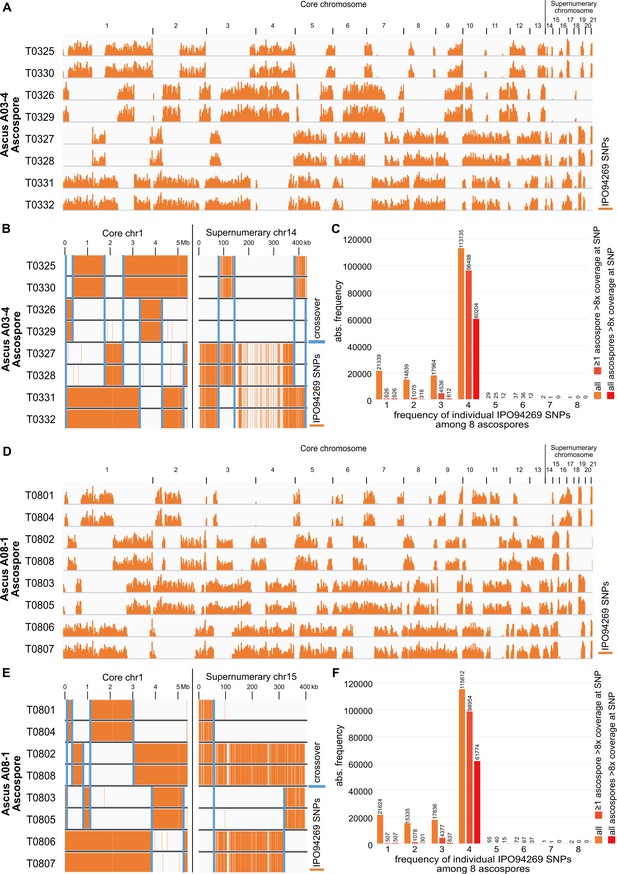
Distribution of SNPs on core and supernumerary chromosomes in two fully sequenced tetrad.
The distribution is consistent with the origin of each tetrad in one meiotic event and Mendelian segregation for core and paired supernumerary chromosomes. (A) Distribution of IPO94269 specific SNP in the eight ascospores of ascus A03-4 mapped on the IPO323 reference genome. Stretches of IPO94269 haplotype alternate with absence of IPO94269 specific SNPs. (B) Detail of distribution of IPO94269 specific SNPs exemplified on core chromosome one and paired supernumerary chromosome 14 of the eight ascospores of ascus A03-4. Recombination events leading to crossover are indicated by blue bars. A total of seven and four crossover involving all sister-chromatids did occur on the core chromosome one and paired supernumerary chromosome 14, respectively. The distribution of the haplotypes is consistent with their origin in one meiotic event. (C) Distribution of the count SNP among the eight ascospores of ascus A03-4 for core and paired supernumerary chromosomes. Mendelian segregation predicts four of the eight ascospores to contain the SNP. Increasing the fidelity of the SNP detetection increased the fraction of SNPs detected in exactly four ascospores. Including all SNP regardless of sequencing coverage in the analsysis results in 68% of the SNPs showing Mendelian segregation. Restricting the analysis to those SNP variants that showed at least in ascospore a sequecning coverage higher than eightfold increased the fraction of SNPs that showed Mendelian segregation to 94%. Further increasing the fidelity of the SNP detection by restricting the analysis to those SNPs that were always detected at a eightfold coverage further increased the fraction of SNPs that showed Mendelian segregation to 97% (ascus A03-4) or 98% (ascus A08-1). (D–F) Distribution of the SNPs of core and supernumerary chromosomes in the eight ascospores of the ascus A08-1, analog to A-C).
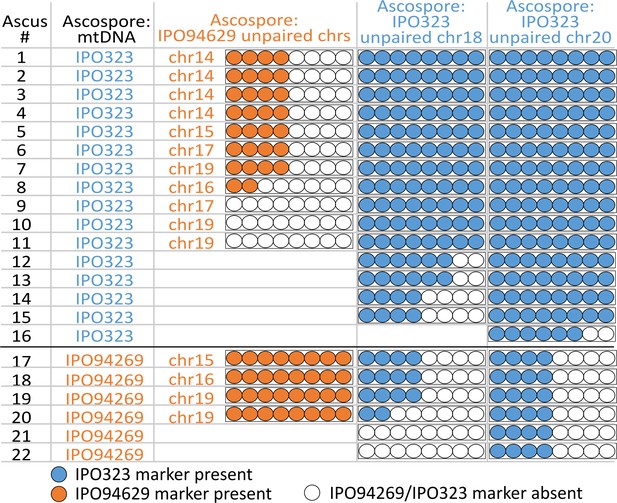
Unpaired supernumerary chromosome show meiotic drive if inherited from the female parent.
Analysis of segregation of unpaired supernumerary chromosomes in 24 complete tetrads according to the mitochondrial genotype. The transmission of chromosomes unique to one of the parental strains and the mitochondrial genotype was detected using chromosomal or mitochondrial markers originating from IPO94269 (orange) and IPO323 (blue) in eight ascospores derived from 24 asci. When IPO323 was the female parent (i.e. the ascospores inherited the mitochondrial genotype of the IPO323 parent) unpaired chromosomes 18 and 20 originating from IPO323 show a strong chromosome drive and are overrepresented in the ascospores. When IPO94269 was the female parent unpaired chromosomes originating from IPO94269 show a strong chromosome drive. Unpaired chromosomes originating from the male parent (i.e. the one not donating the mitochondrial genotype) show Mendelian segregation pattern or are lost. Note: for crosses/chromosome combinations where no unpaired chromosome was present no ascus is shown, which reduces the number of shown asci from the 24 asci that were analyzed in both Figure 3 and Figure 4.
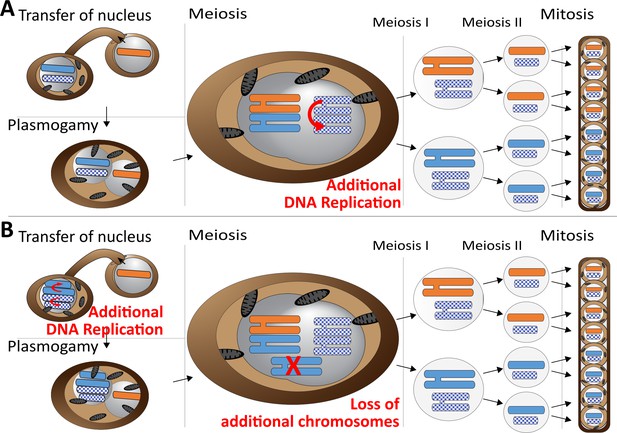
Meiotic chromosome drive in Z.tritici.
Schematic illustration depicting two possible mechanisms for the observed meiotic chromosome drive of female derived unpaired supernumerary chromosomes. Light blue/orange: paired supernumerary chromosome. Checkered blue: unpaired supernumerary chromosome. (A) Chromosome drive occurring during meiosis. Only those unpaired supernumerary chromosomes in the zygote that originated from the female parent are subject to an additional round of DNA replication, allowing for pairing of the two copies of the chromosome. (B) Alternative scenario under which the chromosome drive occurs prior to meiosis. All supernumerary chromosomes are amplified to double the copy number during development of the female ascogonium. The supernumerary chromosomes are paired in the zygote during meiosis. Only additional copies of the supernumerary chromosomes inherited from the female are lost while the supernumerary chromosomes inherited from the male are unaffected.
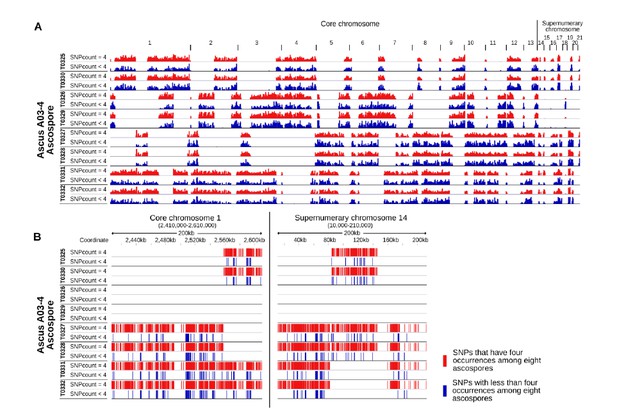
IPO94269 specific SNPs with less than four occurrences among the eight ascospores are interspersed among IPO94269 specific SNPs with exactly four occurrences among the eight ascospores.
A) Whole genome view of all eight ascopsores of ascus A03-4. Stretches of IPO94269 haplotype alternate with stretches of IPO323 haplotype. Crossover events are discernible between all eight ascospores. The distribution of IPO94269 specific SNPs with less than four occurrences among the eight ascospores (blue) is similar to the distribution of the SNPs with exactly four occurrences among the eight ascospores (red). There is no separate clustering of the SNPs showing non-Mendelian inheritance. B) Detailed view of the distribution of SNPs within a representative 200 kb of core chromosome 1 and paired supernumerary chromosome 14 of all eight ascospores of ascus A03-4. IPO94269 specific SNPs with less than four occurrences among the eight ascospores are interspersed among those IPO94269 specific SNPs that show present in four of the eight ascospores.
Tables
Summary of crosses and progeny generated in this study.
https://doi.org/10.7554/eLife.40251.012| Ascospores (random*/all) | Verified tetrads (mtIPO323/mtIPO94269) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| # | Parental strain 1 | Parental strain 2 | Unpaired chr from IPO94269 | Unpaired chr from IPO323 | Condition | Exp A | Exp B | Exp C | Exp A | Exp B | Exp C |
| 1 | IPO323 | IPO94269 | - | chr18, chr20 | Co-inoculation 107 cells/mL | 96/96 | 12/96 | 51/88 | - | 2/2 | 0 |
| 2 | IPO323 ∆chr14 | IPO94269 | chr14 | chr18, chr20 | Co-inoculation 107 cells/mL | 96/96 | 8/64 | 38/72 | - | 3/0 | 1/0 |
| 3 | IPO323 ∆chr21 | IPO94269 | chr21 | chr18, chr20 | Co-inoculation 107 cells/mL | 89/89 | 4/32 | 52/78 | - | 0 | 0 |
| 4 | IPO323 ∆chr16 | IPO94269 | chr16 | chr18, chr20 | Co-inoculation 107 cells/mL | 96/96 | 6/48 | 38/115 | - | 1/0 | 0/1 |
| 5 | IPO323 ∆chr17 | IPO94269 | chr17 | chr18, chr20 | Co-inoculation 107 cells/mL | 96/96 | 2/16 | 15/59 | - | 0 | 2/0 |
| 5 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | Co-inoculation 107 cells/mL | 96/96 | 9/72 | 38/77 | - | 3/1 | 0/1 |
| 7 | IPO323 ∆chr20 | IPO94269 | - | chr18 | Co-inoculation 107 cells/mL | 96/96 | 4/32 | 19/31 | - | 2/0 | 0 |
| 8 | IPO323 ∆chr18 | IPO94269 | - | chr20 | Co-inoculation 107 cells/mL | 96/96 | 4/32 | 30/67 | - | 1/0 | 2/0 |
| 9 | IPO323 ∆chr15 | IPO94269 | chr15 | chr18, chr20 | Co-inoculation 107 cells/mL | 96/96 | 6/48 | 14/54 | - | 1/0 | 0/1 |
| 10 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO323 106 cells/mL | - | - | 25/42 | - | - | 0 |
| 11 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO323 105 cells/mL | - | - | 38/43 | - | - | 0 |
| 12 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO323 104 cells/mL | - | - | 2/16 | - | - | 0 |
| 13 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO94269 +6dpi | - | - | 40/46 | - | - | 0 |
| 14 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO94269 +12dpi | - | - | 11/11 | - | - | 0 |
| 15 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO94269 106 cells/mL | - | - | 60/84 | - | - | 0 |
| 16 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO94269 105 cells/mL | - | - | 48/80 | - | - | 0 |
| 17 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO94269 104 cells/mL | - | - | 28/28 | - | - | 0 |
| 18 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO323 +6pdi | - | - | 5/29 | - | - | 0 |
| 19 | IPO323 ∆chr19 | IPO94269 | chr19 | chr18, chr20 | IPO323 +12dpi | - | - | 51/71 | - | - | 1/0 |
| ∑ | 761/761 | 55/440 | 603/1091 | - | 13/3 | 5/3 | |||||
-
*Includes random and randomized ascospores. Randomized ascospores were generated by randomly selecting one ascospore per tetrad.
Overview of six segregating markers located on the essential chromosomes.
https://doi.org/10.7554/eLife.40251.013| Marker | Localization (IPO323) | Localization (IPO94269) | Primer1 | Primer 2 | Product size in IPO323 [bp] | Product size in IPO94269 [bp] |
|---|---|---|---|---|---|---|
| mat1-1/mat1-2 | chr13 621930–622924 | Unitig11 627136–627792 | MAT1-1F, MAT1-1R | MAT1-2F, MAT1-2R | 340 | 660 |
| 11O21 | chr4 642791–642996 | Unitig03 757794–757990 | 11O21F | 11O21R | 205 | 199 |
| 04L20 | chr4 2276298–2276497 | Unitig03 2427163–2427370 | 04L20F | 04L20R | 192 | 199 |
| caa-0002 | chr3 2927294–2927704 | Unitig02 566646–567041 | 2996 | 2997 | 412 | 396 |
| ggc-001 | chr5 1190388–1190640 | Unitig04 1143568–1143802 | 2998 | 2999 | 254 | 234 |
| ac-0001 | chr7 266661–266847 | Unitiq05 372979–373151 | 3000 | 3001 | 187 | 173 |
Additional files
-
Supplementary file 1
All primers used in this study.
- https://doi.org/10.7554/eLife.40251.014
-
Supplementary file 2
Summary of all PCR marker results for experiment A.
- https://doi.org/10.7554/eLife.40251.015
-
Supplementary file 3
Summary of all PCR marker results for experiment B.
- https://doi.org/10.7554/eLife.40251.016
-
Supplementary file 4
Summary of all PCR marker results for experiment C.
- https://doi.org/10.7554/eLife.40251.017
-
Supplementary file 5
Summary of statistical tests performed in this study.
- https://doi.org/10.7554/eLife.40251.018
-
Supplementary file 6
Frequency of transmission of paired chromosomes to progeny ascospores.
- https://doi.org/10.7554/eLife.40251.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40251.020





