Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway
Figures
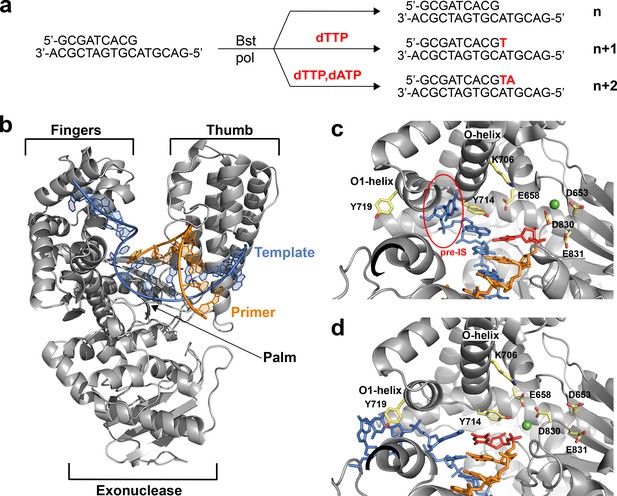
The translocation complex of Bst DNAP-I.
(a) Schematic illustration of the primer-extension reactions used to generate enzyme complexes for the starting duplex (n) and translocated products of the n + 1 and n + 2 nucleotide addition steps. (b) Global architecture of Bst DNAP-I bound to the primer-template duplex (n, 6DSW). (c) The active site region of a known n + 1 in crystallo catalysis structure (1L3T). The pre-insertion site (pre-IS) is circled in red. (d) The active site region of the n + 1 solution-catalyzed reaction (6DSY). Color scheme: polymerase (grey), template (blue), primer (orange), magnesium ion (green), n + 1 nucleotide adduct (red), and amino acid side chains (color by atom).
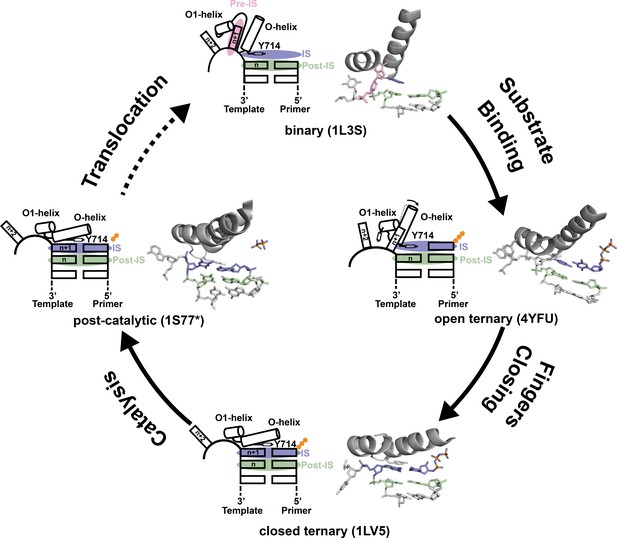
Prevailing mechanism of DNA synthesis by DNA polymerase I.
The four key mechanistic steps of DNA polymerase I have been determined from the structures of Bst DNA polymerase and T7 RNA polymerase (structural homolog). Starting from the binary complex, the n + 1 templating base resides in the pre-insertion site (pre-IS, pink), located in a hydrophic pocket formed by the O-O1 helices and Tyr714 occupies the insertion site (IS, purple), stacking above the newly formed base pair located in the post insertion site (post-IS, green). Upon dNTP binding, the O-O1 helices undergo a minor conformational change, which displaces the n + 1 base from the pre-IS to produce a ternary complex with the incoming dNTP substrate pairing opposite Tyr714 in the IS. The pre-catalytic state is defined by a more significant conformation change where the O-O1 helices close to allow the formation of a nascent base pair between the n + 1 base and incoming dNTP substrate. Following catalysis, the O-O1 helices remain closed with the displaced pyrophosphate moiety observed as a trapped intermediate. Structural details that occur between post-catalysis, translocation, and formation of the subsequent binary complex remain poorly understood (dotted line).
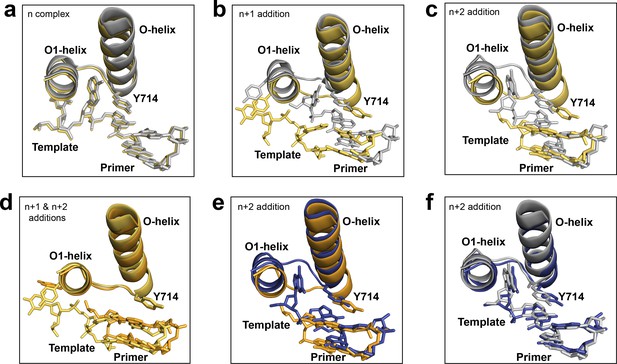
Conformational changes at the active site of Bst DNAP-I.
(a) Crystal structures of the starting primer-template complex (n). Color scheme and PDB codes: new structure (yellow, 6DSW), known structure (grey, 1L3S). (b–f) Post-chemistry structures comparing the active site conformation for n + 1 and n + 2 translocated products generated from solution and in crystallo catalyzed primer-extension reactions. Color scheme and PDB codes: known n + 1 and n + 2 in crystallo catalyzed structures (grey, 1L3T and 1L3U, respectively), new n + 1 and n + 2 solution-catalyzed structures (yellow, 6DSY and 6DSV, respectively), and a new n + 2 in crystallo catalyzed structure obtained from the n + 1 crystal of a solution-catalyzed reaction (blue, 6DSX). (b,c) Overlays comparing the translocated product obtained from solution and in crystallo catalyzed n + 1 and n + 2 primer-extension reactions, respectively. (d) Overlays of the n + 1 and n + 2 structures from solution catalyzed reactions. (e) Overlay of the n + 2 structures for solution and in crystallo catalyzed reactions that initiate from the same polymerase conformation. (f) Overlay of n + 2 in crystallo catalyzed structures that initiate from different polymerase conformations.
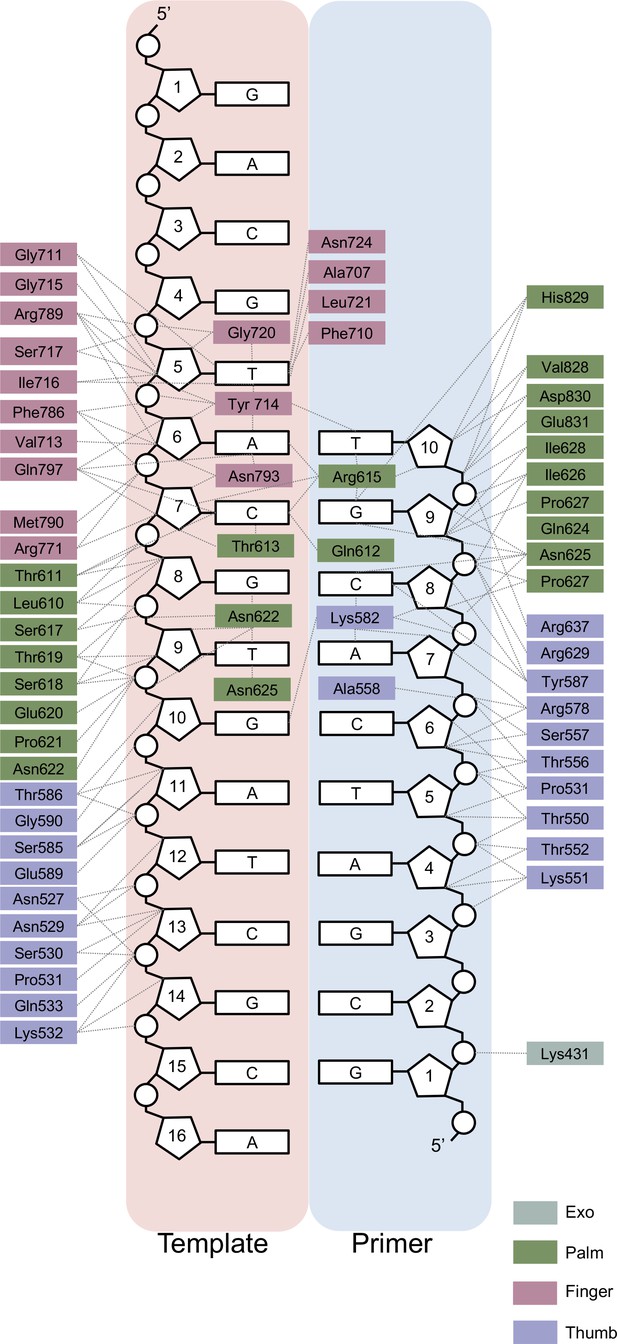
Two-dimensional interaction map for the n + 1 structure obtained from in crystallo catalysis.
Amino acid residues are depicted as boxes, color-coded by polymerase sub-domain. Thin dashed lines represent interactions between the polymerase and a component of the primer/template duplex.
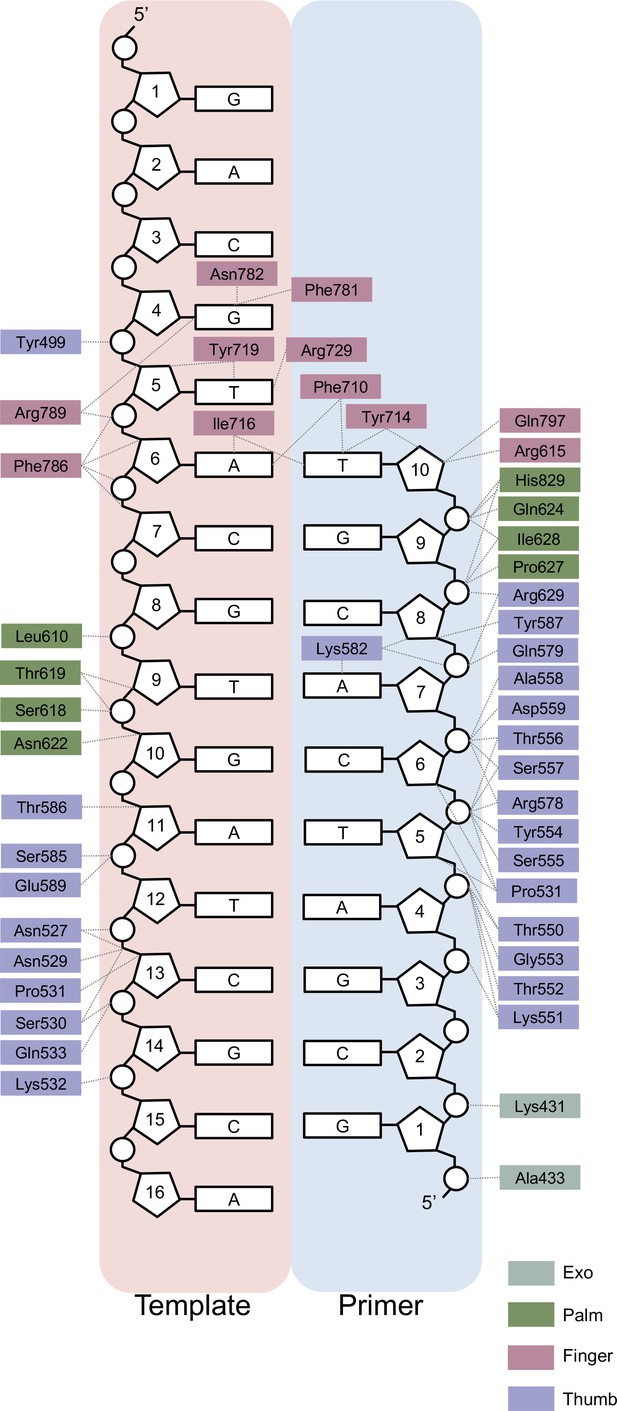
Two-dimensional interaction map for n + 1 structure obtained from a solution-catalyzed reaction.
Amino acid residues are depicted as boxes, color-coded by polymerase sub-domain. Thin dashed lines represent interactions between the polymerase and a component of the primer/template duplex.
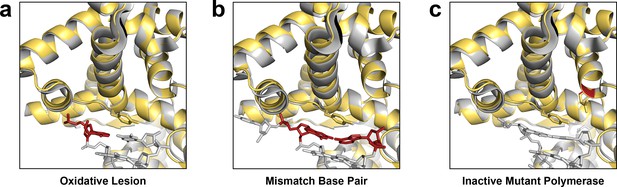
Crystal structures obtained from solution catalyzed reactions resemble active site conformations observed in ‘distorted’ conformations.
Crystal structures obtained by solution catalysis (yellow) are conformationally identical to known binary structures (grey) containing (a) an oxidative lesion at the active site (4B9M), (b) an A:G mismatched in the active site (1NK0), and (c) an inactive Bst mutant (4E0D). Unnatural modifications are highlighted in red.
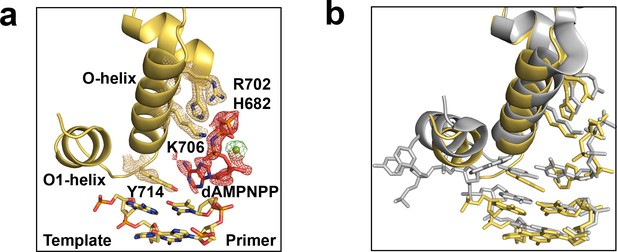
The pre-insertion complex of Bst DNAP-I.
(a) An open ternary structure of Bst DNAP-I (yellow) with a primer-template duplex (color by atom), non-hydrolyzable dATP analog (dAMPNPP, red), and magnesium ion (green) bound in the enzyme active site. Superimposed on the stick model is a 2Fo-Fc omit map contoured at 2.0σ for interacting residues, yellow mesh, and Fo-Fc omit maps contoured at 1.0σ for dAMPNPP and magnesium, red and green mesh, respectively. (b) Comparison of the new ternary structure (yellow, 6DSU) superimposed on a mutant Bst DNAP-I structure solved with dATP bound in the enzyme active site (grey, 4YFU).
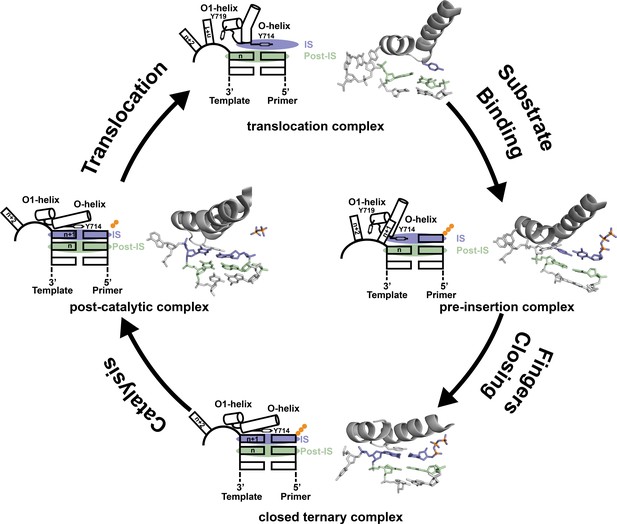
Revised mechanism of DNAP-I.
The four key mechanistic steps of DNAP-I depict a replication cycle for DNA synthesis. The translocation complex (top) is stabilized by π-stacking interactions between Tyr719 and the n + 1 templating base and between Tyr714 and the primer strand. Tyr714 occupies the insertion site (IS, purple) while a newly formed base pair is located in the post insertion site (post-IS, green). In the pre-insertion complex (right), the O-helix adjusts to accommodate the incoming dNTP substrate, which binds opposite Tyr714 in the IS. In the closed ternary complex (bottom), the polymerase undergoes a major conformational change to allow the n + 1 templating base to form a nascent base pair with the dNTP substrate in pre-catalytic state. Following catalysis, the finger subdomain remains closed with a trapped pyrophosphate moiety observed in the active site of the post-catalytic complex (left). To complete the cycle, the finger subdomain opens, pyrophosphate is released, and the enzyme translocates to the next position on the template. The translocation (6DSY), pre-insertion (6DSU), and closed ternary complexes (1VL5) are based on crystal structures Bst DNAP-I. The post-catalytic complex is based on the structure of T7 RNAP (1S77), which is a homolog of Bst DNAP-I.
Tables
Data collection and refinement statistics
https://doi.org/10.7554/eLife.40444.009| N | n + 1 | n + 1, dATP soak | n + 1, dAMPNPP soak | n + 2 | |
|---|---|---|---|---|---|
| Data Collection | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell Dimensions | |||||
| a, b, c (Å) | 86.1, 93.4, 105.6 | 88.1, 93.7, 105.8 | 87.1, 93.5, 105.3 | 87.44, 93.39, 104.95 | 87.0, 93.0, 104.7 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 54.31–1.58 (1.64–1.58) | 46.09–1.98 (2.05–1.98) | 46.7–1.99 (2.06–1.99) | 43.72–1.74 (1.78–1.74) | 41.04–1.99 (2.06–1.99) |
| Rmerge | 0.7309 (1.35) | 0.7085 (1.389) | 0.1329 (0.7194) | 0.0567 (0.241) | 0.3279 (1.745) |
| CC1/2 | 0.842 (0.795) | 0.759 (0.543) | 0.993 (0.796) | 0.999 (0.977) | 0.991 (0.586) |
| I / σI | 71.43 (3.56) | 43.97 (2.64) | 17.03 (2.90) | 22.78 (9.66) | 9.75 (2.49) |
| Completeness (%) | 99.98 (99.98) | 96.97 (99.15) | 99.92 (99.90) | 98.25 (99.33) | 99.90 (99.93) |
| Redundancy | 31.3 (25.0) | 12.9 (11.0) | 6.8 (4.7) | 7.0 (6.8) | 7.2 (7.3) |
| Refinement | |||||
| Resolution (Å) | 54.31–1.58 (1.64–1.58) | 46.09–1.98 (2.05–1.98) | 46.7–1.99 (2.06–1.99) | 43.72–1.98 (2.05–1.98) | 41.04–1.99 (2.06–1.99) |
| No. reflections | 115039 (11360) | 59886 (6056) | 59677 (5857) | 59416 (5901) | 58990 (5831) |
| Rwork/Rfree | 0.165/0.189 (0.199/0.248) | 0.202/0.255 (0.264/0.340) | 0.184/0.219 (0.239/0.293) | 0.222/0.271 (0.225/0.281) | 0.192/0.228 (0.332/0.391) |
| No. atoms | 5961 | 4627 | 5412 | 5468 | 5453 |
| Protein | 4636 | 4627 | 4639 | 4661 | 4590 |
| Duplex | 490 | 469 | 487 | 429 | 475 |
| Solvent | 835 | 546 | 286 | 378 | 388 |
| B-factors | 26.73 | 42.07 | 45.96 | 42.25 | 39.05 |
| Protein | 25.11 | 42.17 | 45.39 | 41.74 | 38.85 |
| Duplex/dAMPNPP | 40.11 | 55.17 | 117.16 | 101.56/106.4 | 60.07 |
| Solvent | 36.64 | 40.95 | 46.01 | 41.13 | 41.37 |
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.006 | 0.007 | 0.008 | 0.008 | 0.007 |
| Bond angles (°) | 0.82 | 0.89 | 0.84 | 1.15 | 0.85 |
-
*Values in parentheses are for the highest-resolution shell.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | DH5-αderivative | NEB | C2987H | Chemically competent cells for recombinant expression of Bst DNAP-I |
| Recombinant DNA reagent | pDEST007-Bst | PMID: 20813757 | Original expression plasmid for Bst DNAP-I | |
| Recombinant DNA reagent | pGDR11 | PMID: 9401025 | Expression plasmid for Bst DNAP-I used in this study | |
| Sequence-based reagent | Bst_for | IDT | 5’-ATCCATATGGCATTT ACGCTTGCTGAC-3’ | |
| Sequence-based reagent | Bst_rev | IDT | 5’-ATGCGGCGGTCTCC TCGAGTCATTATTT CGCATCATACCACG-3’ | |
| Sequence-based reagent | DNA template | IDT | 5’-GACGTACG TGATCGCA-3’ | |
| Sequence-based reagent | DNA primer | IDT | 5’- GCGATCACGT-3’ | |
| Software, algorithm | XDS | PMID: 20124692 | RRID: SCR_015652 | |
| Software, algorithm | Phaser | PMID: 19461840 | RRID: SCR_014219 | |
| Software, algorithm | Phenix refine | PMID: 22505256 | RRID: SCR_014224 | |
| Software, algorithm | Coot | PMID: 20383002 | RRID: SCR_014222 | |
| Software, algorithm | Molprobity | PMID: 2057044 | RRID: SCR_014226 |
Additional files
-
Supplementary File 1
a.Helical statistics for solution and in crystallo catalyzed n + 1 translocated structures.
b.Structures of Bst DNAP-I with homologous active sites.
- https://doi.org/10.7554/eLife.40444.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40444.013





