FACT and Ubp10 collaborate to modulate H2B deubiquitination and nucleosome dynamics
Figures
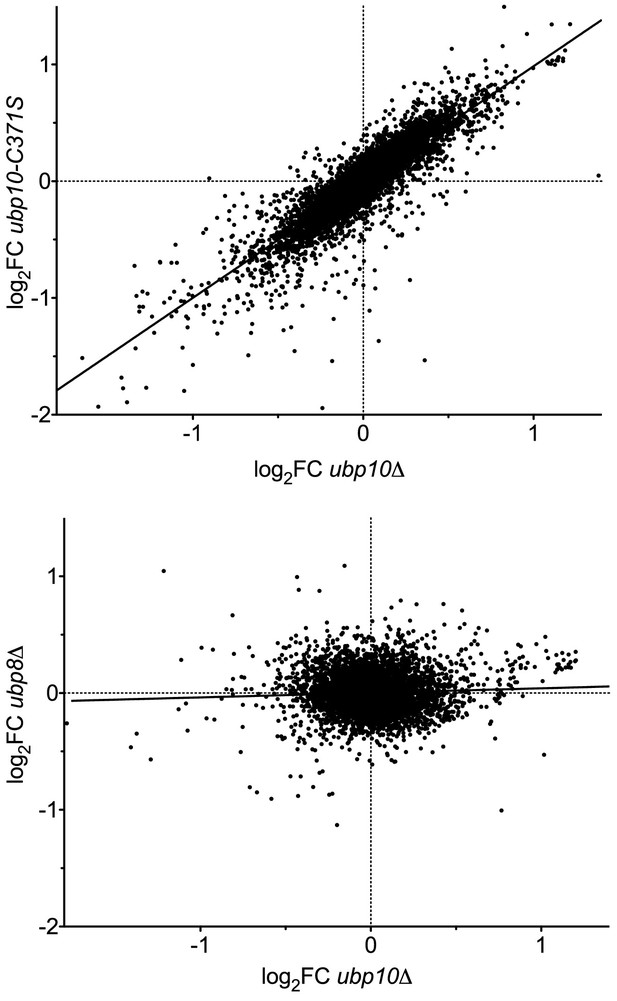
Deletion of the UBP8 and UBP10 genes have different effects on transcription programs.
Analysis of transcription data from Gardner et al., 2005. Scatter plots of the log2 fold change in transcript level relative to WT (log2FC) are shown for (top panel) a catalytically dead allele (ubp10-C371S) vs a deletion (ubp10∆) to demonstrate reproducibility of the array, and (bottom panel) a ubp8∆ strain compared with ubp10∆. The two null mutants give a strong correlation (Pearson correlation coefficient r = 0.86, linear regression R2 = 0.74, m = 0.99), validating the reproducibility of the arrays. Deleting UBP8 affected the transcription of different genes, resulting in poor correlation with ubp10∆ (r = 0.055, R2 = 0.0031, m = 0.039).
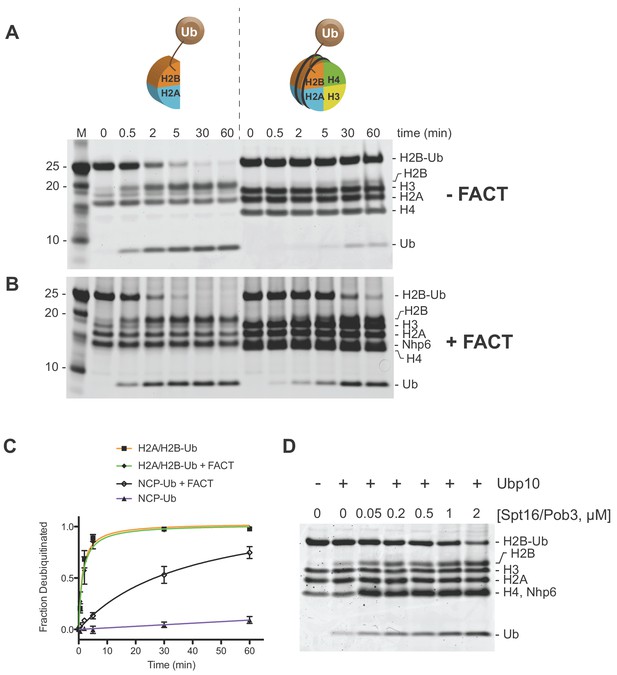
Ubp10 preferentially deubiquitinates H2A/H2B-Ub over nucleosomal (NCP) H2B-Ub.
(A–B) Comparison of Ubp10 activity on H2A/H2B-Ub and NCP-Ub in the absence (A) and presence of FACT (B). In panel A, 1 µM NCP-Ub and 2 µM H2A/H2B-Ub were incubated with 5 nM Ubp10 and time points were taken by quenching with SDS sample buffer. (B) FACT stimulates Ubp10 activity on NCP-Ub. Ubp10 activity was measured as in A, but in the presence of FACT subunits, 2 µM Spt16/Pob3-WT and 2 µM Nhp6. (C) The fraction of total substrate consumed over time from assays performed in A-B is shown. The plot was generated by averaging the relative intensity of H2B-Ub bands as compared with uncleaved H2B-Ub at t = 0 from three independent experiments (mean normalized band intensity and standard deviation shown). (D) Increasing the concentration of FACT increases the activity of Ubp10. Enzyme activity was monitored by mixing 1 µM NCP-Ub, 2 µM Nhp6, and the indicated concentrations of Spt16/Pob3 in the presence of 5 nM Ubp10. Each reaction was quenched at 60 min.
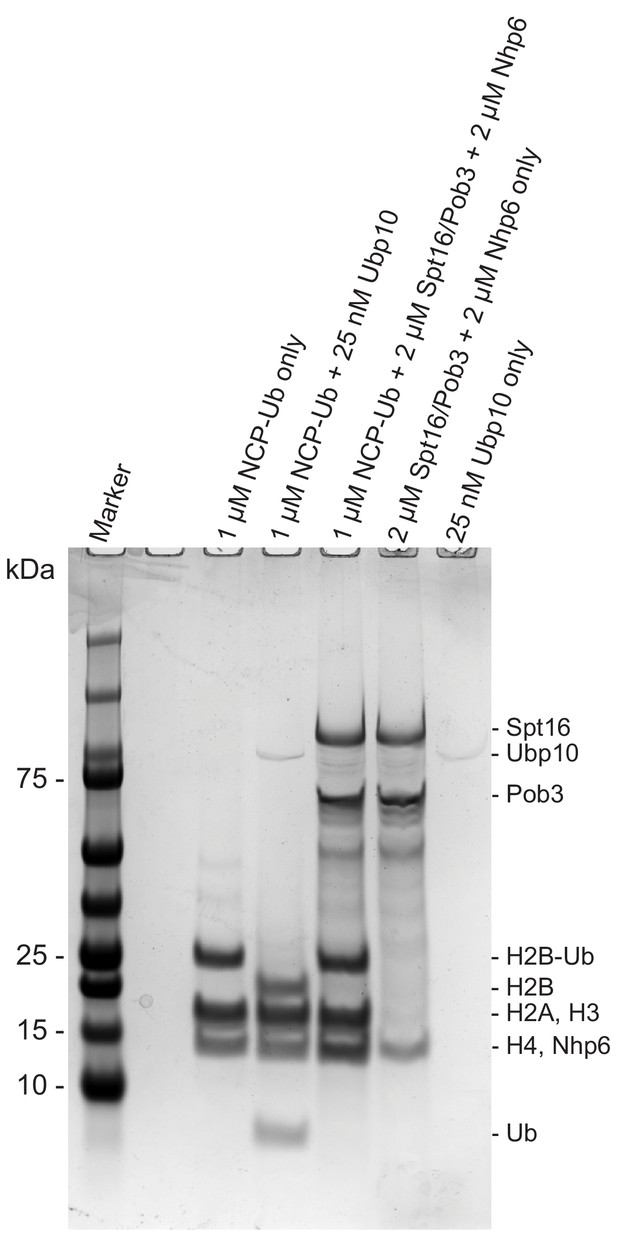
FACT alone does not deubiquitinate H2B-Ub.
Coomassie-stained gel of a control experiment demonstrating that purified Spt16/Pob3 and Nhp6 do not contain contaminants that deubiquitinate H2B-Ub. Proteins indicated in each lane were incubated for 3 hr after which the reactions were quenched.
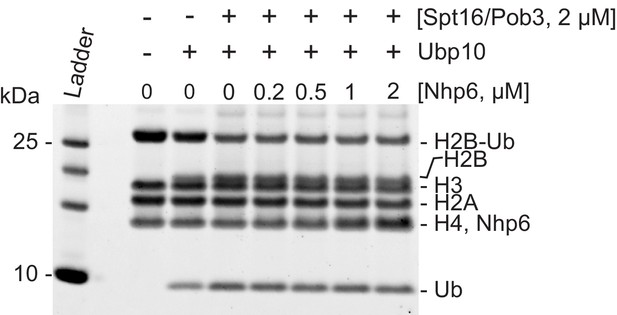
FACT does not require Nhp6 to stimulate H2B deubiquitination.
A deubiquitination assay was performed as in Figure 2D, except with variable concentrations of Nhp6. Spt16/Pob3 enhanced the deubiquitinase activity but adding Nhp6 did not enhance this effect noticeably.
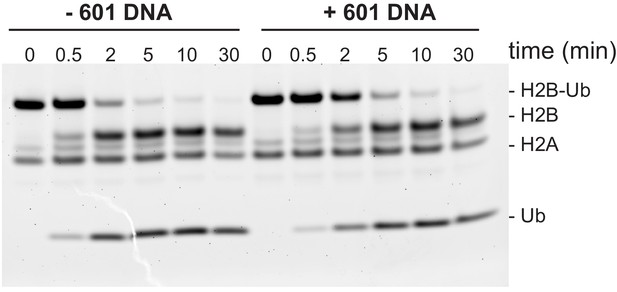
Ubp10 activity on ubiquitinated dimers is unaffected by the presence of 601 DNA.
Ubp10 activity on H2A/H2B-Ub with and without 601 DNA. For reaction with 601 DNA, 2 µM of 146 bp Widom 601 DNA was incubated with 2 µM H2A/H2B-Ub for 1 hr at 4° C. Then the reaction was transferred to a 30°C water bath and DUB activity was monitored as in Figure 2.
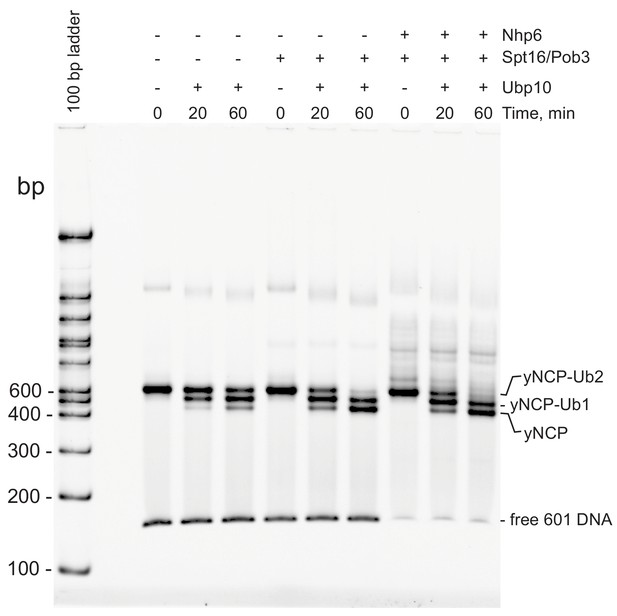
The presence of FACT does not alter nucleosome integrity during H2B deubiquitination.
Deubiqutination was measured as in Figure 2A–B; following incubation at 30°C, the reactions were transferred to ice. Mobility was monitored on a native 6% polyacrylamide gel after diluting samples 6-fold. Note that Spt16/Pob3 stimulates the activity of Ubp10 even in the absence of Nhp6, and that adding Nhp6 does not affect the integrity of the nucleosomal products as judged by their rate of migration through this native gel. The concentration of Nhp6 after dilution is sufficient to bind the free DNA species, and to make transient complexes with nucleosomes, causing loss of the free DNA form and formation of additional bands above the labeled yNCP species, but the yNCP species migrate in the same positions, suggesting that they have the same composition.
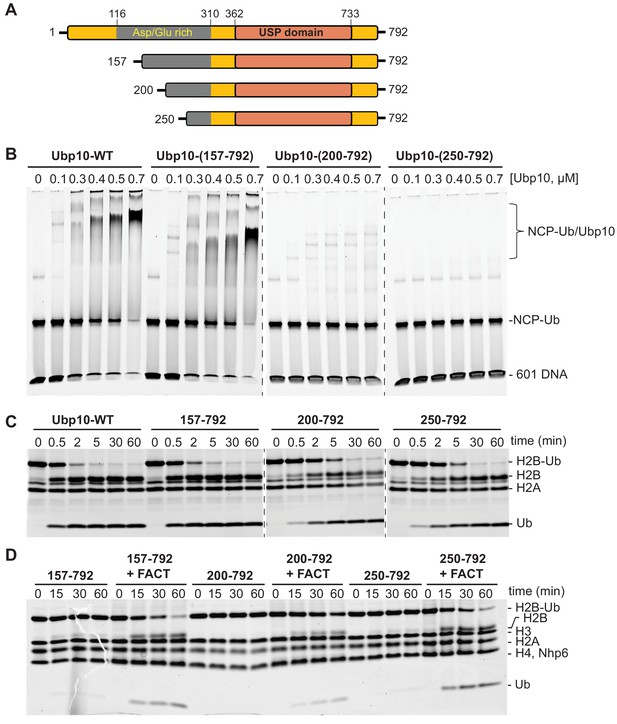
FACT stimulates Ubp10 constructs that lack intrinsic ability to bind nucleosomes.
(A) Schematic of Ubp10 and its truncations with the locations of the catalytic domain (USP) and an N-terminal region rich in aspartic acid and glutamic acid repeats shown. (B) Native gel showing complexes formed between Ubp10 constructs and ubiquitinated nucleosomes (purified proteins used in this experiment are shown in Figure 3—figure supplement 4). (C) Ubp10 activity was measured as in Figure 2; time points were taken after incubating 2 µM H2A/H2B-Ub dimers with 5 nM Ubp10 fragments. (D) NCP-Ub cleavage activity monitored in the presence and absence of FACT and several Ubp10 constructs.
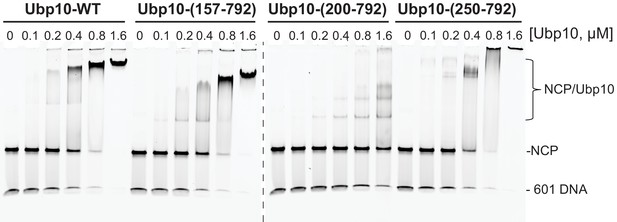
Native gel showing binding of several Ubp10 fragments to unmodified yeast nucleosomes.
Reaction conditions are the same as in Figure 3B.
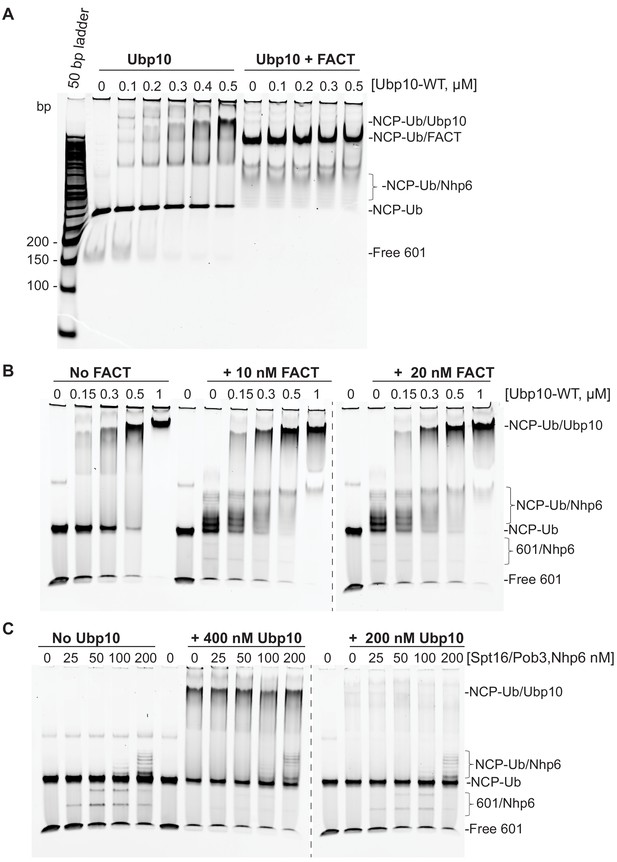
FACT does not affect the affinity of Ubp10 for nucleosomes.
Electrophoretic mobility shift assay (EMSA) of Ubp10 binding to ubiquitinated nucleosomes in the presence and absence of FACT. (A) Ubp10/NCP-Ub binding monitored by incubating 100 nM NCP-Ub in EMSA buffer in the absence (left) or presence (right) of FACT (2 µM Nhp6 and 2 µM Spt16/Pob3-WT) and with increasing concentrations of Ubp10. Samples were incubated for 1 hr on ice and electrophoresed on a 4% native polyacrylamide gel. At high concentrations of FACT, the binding of FACT to nucleosomes blocks the formation of stable Ubp10-nucleosome complexes. (B) Reactions in the presence of 2 µM Nhp6 and 10 nM Spt16/Pob3 (left) or 20 nM Spt16/Pob3 (right). (C) Reactions with equimolar concentrations of FACT subunits at the apparent KD (left) and below the KD of the binding of Ubp10 to NCP-Ub. Samples in B and C were analyzed on a 6% polyacrylamide TBE gels (Life Technologies).
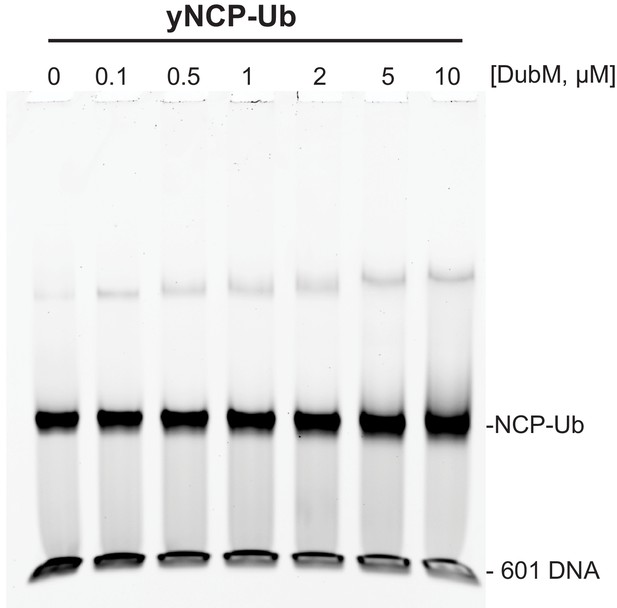
Binding of the SAGA DUB module to ubiquitinated nucleosomes is not detectable by EMSA.
Increasing amounts of DubM were incubated for 1 hr with 100 nM yeast ubiquitinated nucleosomes (DUB resistant yNCP-DCA-Ub, see methods). Following native gel electrophoresis, the gel was stained with SYBR Gold. The mobility of nucleosomes remained unchanged with increasing concentrations of the DUB module.
SDS-PAGE gel showing purity of Ubp10 truncations.
SDS-PAGE gel stained with Coomassie Brilliant Blue showing each of the purified Ubp10 truncations used in Figure 3.
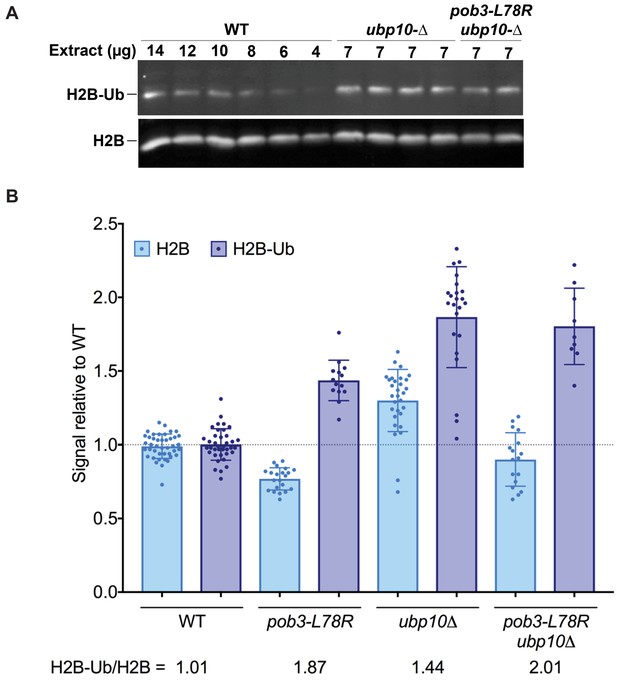
H2B-Ub levels are elevated in a FACT mutant strain.
(A) Representative western blot analysis of TCA extracts from WT, ubp10Δ, and ubp10Δ pob3-L78R strains probed with antibodies against H2B-Ub and re-probed with antibodies against H2B. (B) Relative steady-state levels of H2B-Ub for WT, pob3-L78R, ubp10Δ, and ubp10Δ/pob3-L78R strains (Table 1). The average and standard deviation from multiple biological replicates is shown. The numbers at the bottom indicate the relative H2B-Ub increase normalized to the paired unmodified H2B value (H2B-Ub/H2B) within each individual gel. Total H2B is decreased in the mutant because the slow growth of the strain leads to a larger average cell size and therefore a lower contribution of nuclear proteins to the total protein level that was used to normalize loading.
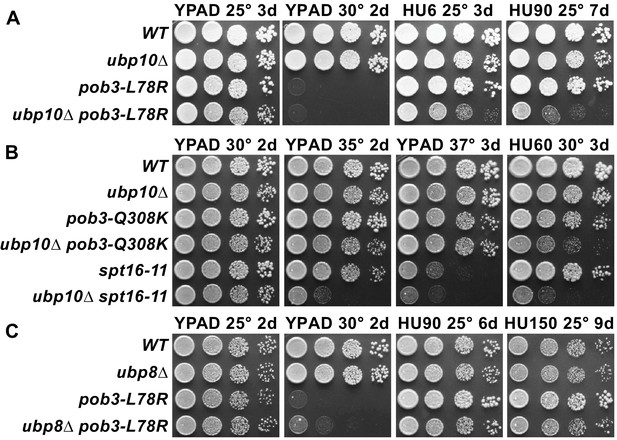
Combining ubp10∆ and FACT alleles causes sensitivity to HU.
(A–C) Strains indicated (Table 1) were grown to saturation, then 10-fold serial dilutions were spotted on rich medium (YPAD) with and without the indicated concentrations of hydroxyurea (HU, mM). Plates were incubated at the temperature indicated for the time shown (days). Combining pob3-L78R with ubp10∆ caused HU sensitivity (A), but combining it with ubp8∆ did not (C). Combining ubp10∆ with other alleles of FACT also caused synthetic defects on HU (B).
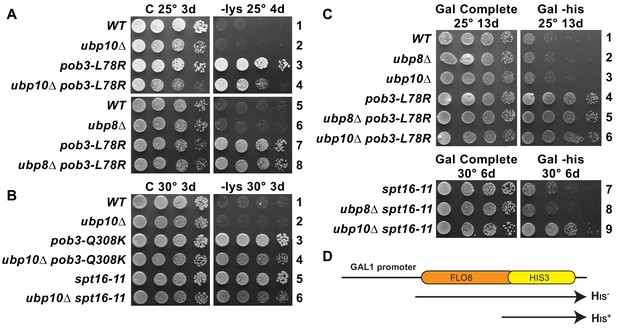
Combined effect of FACT mutants and ubp10∆ or ubp8∆ deletions on the Spt– phenotype and cryptic promoter activation.
(A,B) ubp10∆ did not affect the Spt– phenotype of FACT mutants. Dilutions of the same strains shown in Figure 5 were plated on synthetic medium with (C, complete) or without lysine (-lys). These strains with the lys2-128∂ allele are auxotrophic for lysine, but defects in chromatin integrity allow expression of the gene, which is revealed as growth on -lys (the Spt– phenotype; see Simchen et al., 1984). FACT mutants displayed this phenotype, but this was not affected by ubp8∆ or ubp10∆ (A, B, and not shown). (C) Activation of a cryptic transcription reporter in a ubp10∆/spt16-11 mutant strain reveals a defect in restoring chromatin in the wake of RNA Pol II passage. Strains with an out-of-frame fusion of HIS3 to a site downstream of a cryptic promoter in the FLO8 gene (panel D, adapted from Cheung et al., 2008) are auxotrophic for histidine when the GAL1 promoter driving transcription of this reporter is repressed on glucose (not shown) but can grow without histidine on synthetic medium containing galactose (Gal -his). Strains with the pob3-L78R mutation have this phenotype, indicating activation of the cryptic promoter, masking any potential effects of ubp8∆ or ubp10∆. The spt16-11 allele alone did not activate this reporter but did when combined with ubp10∆.
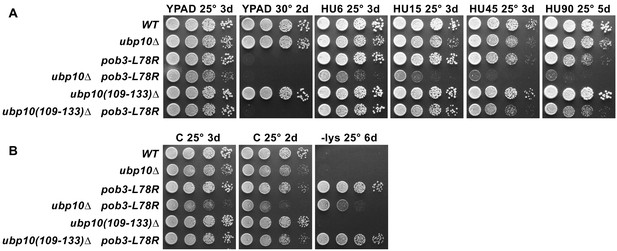
Removing the SIR-interaction domain (residues 109–133) from Ubp10 does not affect the phenotypes of a FACT mutant.
(A) Strains indicated (Table 1) were grown to saturation, then 10-fold serial dilutions were spotted on rich medium (YPAD) with and without the indicated concentrations of hydroxyurea (HU, mM). Plates were incubated at the temperature indicated for the time shown (days). Combining pob3-L78R with ubp10(109-133)∆ does not cause the synthetic growth defects seen in the full ubp10 deletion. (B) Genetic analysis as in Figure 6A–B, but with the ubp10 internal deletion lacking the SIR-complex interaction domain. ubp10(109-133)∆ does not cause the growth defect observed with ubp10∆, and neither affect the Spt- phenotype (the weaker growth of the pob3-L78R/ubp10∆ strain on -lys parallels its weaker growth on rich media; see Figure 6A). Together, these results show that the phenotypes we observe in the pob3-L78R/ubp10∆ strain (Figures 5–6) are not caused by failure of Ubp10's role in silencing.
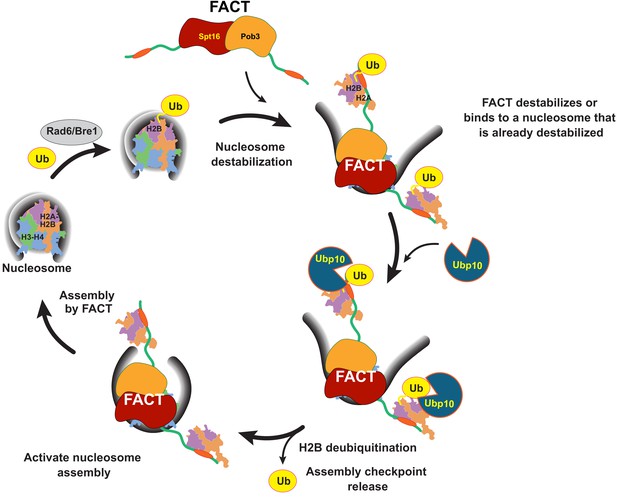
Model for coordinated H2B deubiquitination and nucleosome assembly by FACT/Ubp10 during transcription and DNA replication.
Rad6/Bre1 ubiquitinates nucleosomal histone H2B during transcription and DNA replication. The presence of H2B-Ub recruits FACT to the nucleosome. FACT destabilizes nucleosomes or binds to nucleosomes that are already destabilized by transcription or replication factors. Ubp10 deubiquitinates H2B-Ub in the context of fully or partially exposed H2A/H2B-Ub heterodimers while still tethered to Spt16/Pob3. Deubiquitination of H2B signals passage of polymerases and deposition of histones in the wake of polymerases. The deubiquitinated nucleosome is reassembled by FACT, followed by dissociation of FACT. The full reorganization depicted here supports Ubp10 activity, but since Spt16-Pob3 heterodimers can support Ubp10 as well, activation of Ubp10 may require only the early stages of reorganization that are not dependent on Nhp6.
Tables
Yeast Strains used All strains are congenic with the A364a background and are MATa.
Standard methods were used to introduce the mutations shown into diploid strains, then haploids were derived and crossed to obtain the combinations listed, ensuring that all strains with the same genotype displayed the phenotypes observed.
| Figure 4 Western blots | ||
|---|---|---|
| Strain | Label | Genotype |
| 8127-7-4 | WT | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ |
| 10018-1-4 | ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::KanMX) |
| 9204 | pob3-L78R | ura3 leu2 trp1 his3 lys2-128∂ pob3-L78R(+34, LEU2) |
| 10025-2-4 | pob3-L78R ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::HphMX) pob3-L78R(+34, LEU2) |
| Figure 5A (top panel), Figure 6A (1-4) | ||
| 8127-7-4 | WT | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ |
| 10018-1-4 | ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::KanMX) |
| 9204 | pob3-L78R | ura3 leu2 trp1 his3 lys2-128∂ pob3-L78R(+34, LEU2) |
| 10025-2-4 | pob3-L78R ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::HphMX) pob3-L78R(+34, LEU2) |
| Figure 5C (middle panel), Figure 6A (5-8) | ||
| 8127-7-4 | WT | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ |
| 8540-1-1 | ubp8∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp8-∆(::KanMX) |
| 9204 | pob3-L78R | ura3 leu2 trp1 his3 lys2-128∂ pob3-L78R(+34, LEU2) |
| 10032-4-3 | pob3-L78R ubp8∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ pob3-L78R(+34, LEU2) ubp8-∆(::KanMX) |
| Figure 5B (bottom panel), Figure 6B | ||
| 8127-7-4 | WT | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ |
| 10018-1-4 | ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::KanMX) |
| 9273H | pob3-Q308K | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ pob3-Q308K(+34, HphMX) |
| 10019-2-3 | pob3-Q308K ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ pob3-Q308K(+34, HphMX) ubp10-∆(::KanMX) |
| 9495 H-2-3 | spt16-11 | ura3 leu2 trp1 his3 lys2-128∂ spt16-11(+124, HphMX) |
| Figure 6C | ||
| 9880-2-2 | WT | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) |
| 10040-3-2 | ubp8∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) ubp8-∆(::KanMX) |
| 10024-3-1 | ubp10∆ | ura3 leu2-∆0 trp1 his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) ubp10-∆(::HphMX) |
| 10040-1-3 | pob3-L78R | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) pob3-L78R(+34, LEU2) |
| 10040-5-1 | pob3-L78R ubp8∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) pob3-L78R(+34, LEU2) ubp8-∆(::KanMX) |
| 10039-1-4 | pob3-L78R ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) pob3-L78R(+34, LEU2) ubp10-∆(::HphMX) |
| 9949-3-1 | spt16-11 | ura3 leu2 trp1 his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) spt16-11(+124, KanMX) |
| 10044-4-2 | ubp8∆ spt16-11 | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) spt16-11 ubp8-∆(::KanMX) |
| 10043-7-3 | ubp10∆ spt16-11 | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ GAL1pr-flo8-HIS3(NatMX) spt16-11(+124, HphMX) ubp10-∆(::KanMX) |
| 11-16-2018 upb10∆(109-133) tests (Figure 6—figure supplement 1) | ||
|---|---|---|
| 8127-7-4 | WT | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ |
| 10018-1-4 | ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::KanMX) |
| 9204 | pob3-L78R | ura3 leu2 trp1 his3 lys2-128∂ pob3-L78R(+34, LEU2) |
| 10062-4-4 | pob3-L78R ubp10∆ | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(::HphMX) pob3-L78R(+34, LEU2) |
| 10064-2-1 | ubp10∆(109-133) | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ ubp10-∆(109-133) |
| 10062-1-2 | pob3-L78R ubp10∆(109-133) | ura3-∆0 leu2-∆0 trp1-∆two his3 lys2-128∂ pob3-L78R(+34, LEU2) ubp10-∆(109-133) |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40988.019





