The conserved aspartate ring of MCU mediates MICU1 binding and regulation in the mitochondrial calcium uniporter complex
Figures
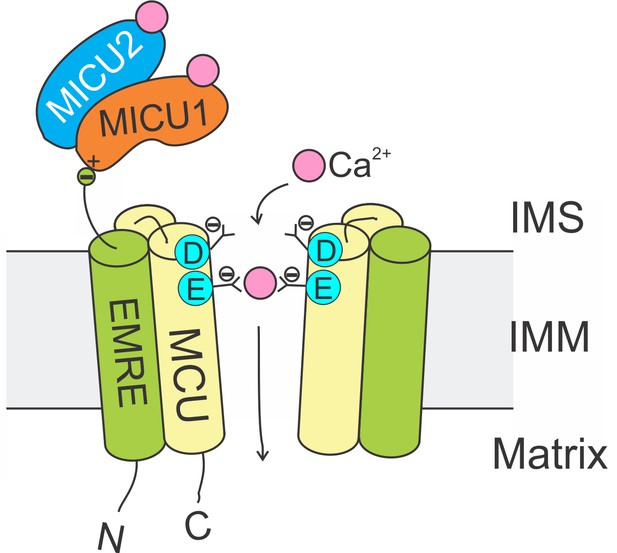
Molecular assembly of the mitochondrial Ca2+ uniporter.
The MCU protein assembles into a tetrameric Ca2+ pathway across the inner mitochondrial membrane (only two subunits are illustrated to reveal the Ca2+ pore). Conserved Asp and Glu residues in MCU’s DIME signature sequence form two parallel side-chain carboxylate rings at the IMS entrance of the pore to coordinate Ca2+. The EMRE protein binds to MCU and MICU1 via its TM helix and C-terminal tail, respectively. When an intracellular Ca2+ signal arrives at the IMS surface of the uniporter, Ca2+ binding to MICUs leads to activation of the uniporter to transport Ca2+ into the matrix.
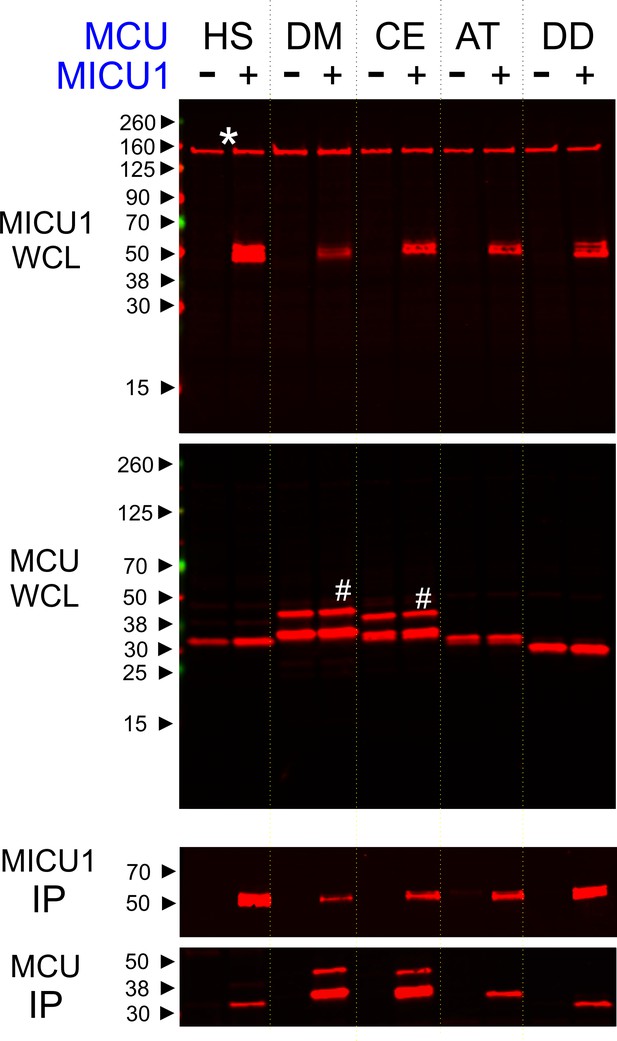
Conserved MCU-MICU1 interactions.
1D4-tagged MCU homologues from various species (HS: Homo sapiens, DM: Drosophila Melanogaster, CE: Caenorhabditis elegans, AT: Arabidopsis thaliana, and DD: Dictyostelium discoideum) were expressed in the presence or absence of FLAG-tagged WT human MICU1 in MCU/EMRE-KO cells. MICU1 was immobilized in FLAG-affinity resins to pull down MCU. Anti-FLAG and anti-1D4 antibodies were used to detect MICU1 and MCU, respectively. SDS-PAGE was performed under reducing conditions. WCL: whole cell lysate. IP: immunoprecipitation. Asterisk: non-specific Western signals. Hash: MCU homologues that contain untruncated mitochondrial-targeting sequences.
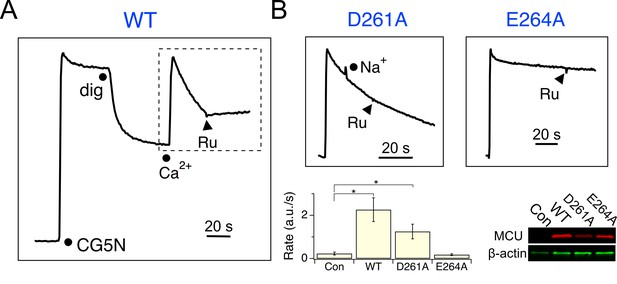
Functional analysis of MCU.
(A) A fluorescence-based mitochondrial Ca2+ uptake assay. MCU-KO HEK293 cells, transiently expressing WT MCU, were permeabilized with digitonin (dig) in the presence of an extracellular Ca2+ indicator Calcium Green-5N (CG5N). Adding 10 µM CaCl2 leads to an immediate increase of fluorescence, followed by a signal decline reflecting uniporter-mediated Ca2+ uptake. Ru360 (Ru) was added to inhibit the channel. In subsequent experiments, only traces obtained after applying Ca2+ (dashed box) are presented. (B) The activity of D261A or E264A mutants. These mutants were expressed in MCU-KO cells, with 100 mM NaCl added during Ca2+ uptake to test if the channel can select Ca2+ against Na+. The bar chart summarizes the initial rate of Ca2+ uptake, and the western blot compares expression levels of MCU constructs. Con: untransfected cells. *p<0.01.
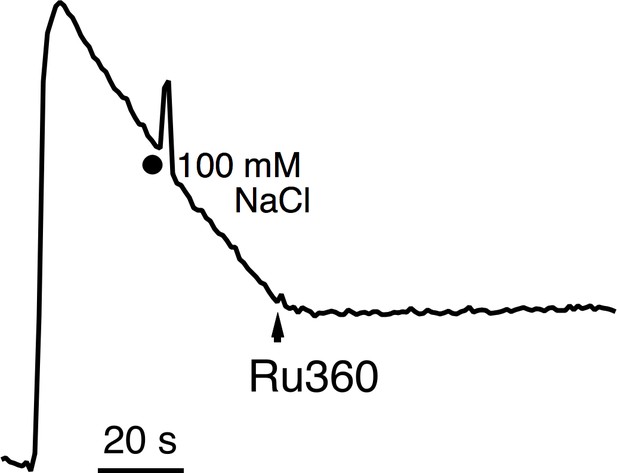
The response of WT MCU to Na+.
100 mM NaCl was added while WT MCU transports Ca2+ (10 µM) into mitochondria.
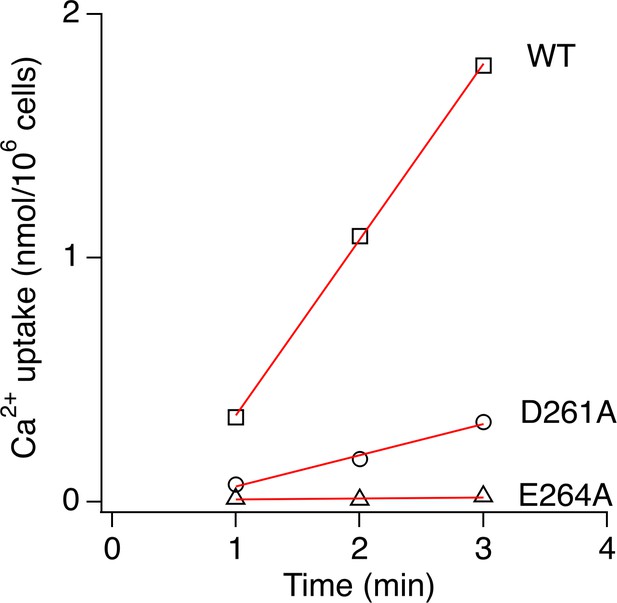
Quantification of uniporter Ca2+ transport.
WT, D261A, or E264A MCU was expressed in MCU-KO cells and their activities were quantified using a 45Ca2+ flux assay in the presence of 10 µM Ca2+. To achieve comparable expression levels of these constructs, we used 1 µg of DNA for WT or E264A and 2.2 µg of DNA for D261A in transfection. In each experiment, 45Ca2+ transported into mitochondria by MCU was measured over a 3 min time course, and the readings were fit with a linear function (red lines) to produce the rate of Ca2+ transport. Rates from three independent experiments were averaged, yielding the following: 610 ± 105 pmol/min/106 cells for WT, 160 ± 23 pmol/min/106 for D261A, and 6 ± 2 pmol/min/106 for E264A.
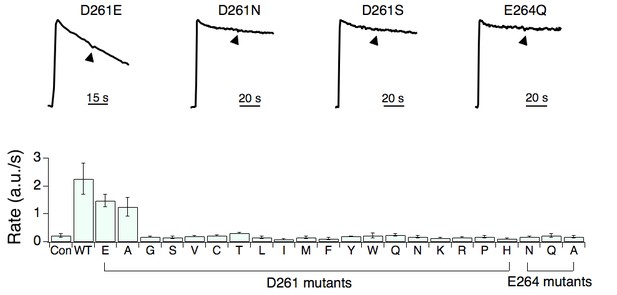
The activity of D261 or E264 MCU mutants.
Various MCU constructs were expressed in MCU-KO cells, and their function was analyzed with a fluorescence-based mitochondrial Ca2+ uptake assay as in Figure 3. Arrowheads indicate 75 nM Ru360. The D261E mutant is functional, but loses sensitivity to Ru360. Con: untransfected MCU-KO cells.
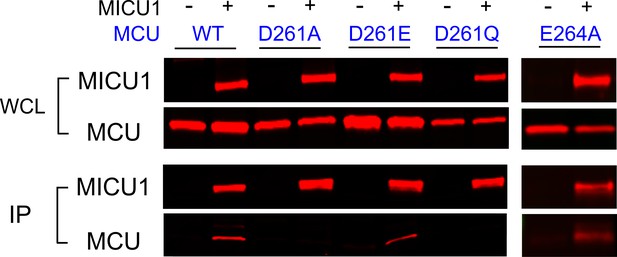
The impact of D261 or E264 mutations on MICU1 binding.
FLAG-tagged WT MICU1 was used to pull down various MCU mutants co-expressed in MCU/EMRE-KO cells.
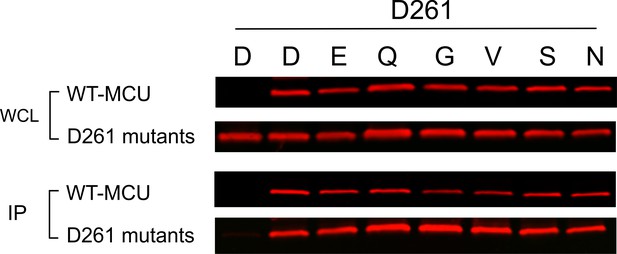
Oligomerization of D261 mutants.
1D4-tagged WT MCU was used to pull down C8 (PRGPDRPEGIEE)-tagged D261 MCU mutants co-expressed in MCU-KO cells. Results show that these D261 mutants complex with WT MCU, suggesting that like WT they assemble into oligomers.
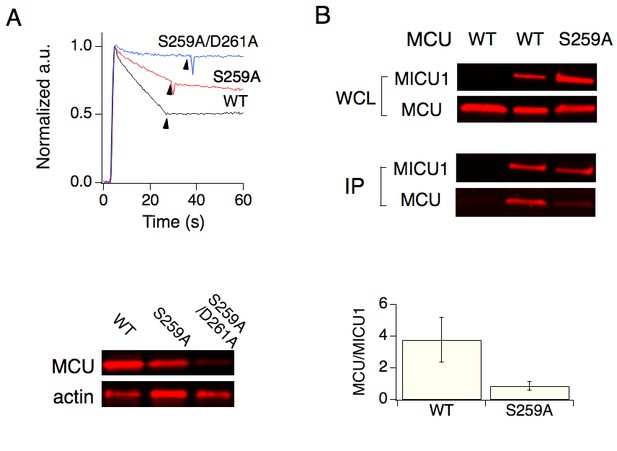
The role of S259 in Ru360 inhibition and MICU1 binding.
(A) The effect of the S259A mutation on mitochondrial Ca2+ transport. WT, S259A, or S259A/D261A MCU was expressed in MCU-KO cells, with activity analyzed using the fluorescence-based Ca2+ flux assay as in Figure 3. Arrowheads mark Ru360 addition. Western images (bottom) show that the S259A/D261A double mutant is poorly expressed. (B) CoIP experiments testing the role of S259 in MICU1 binding. FLAG-tagged MICU1 was used to precipitate 1D4-tagged MCU. The IP signal of MCU was normalized to that of MICU1, as presented in the bar chart. A t-test produces a P value < 0.05.
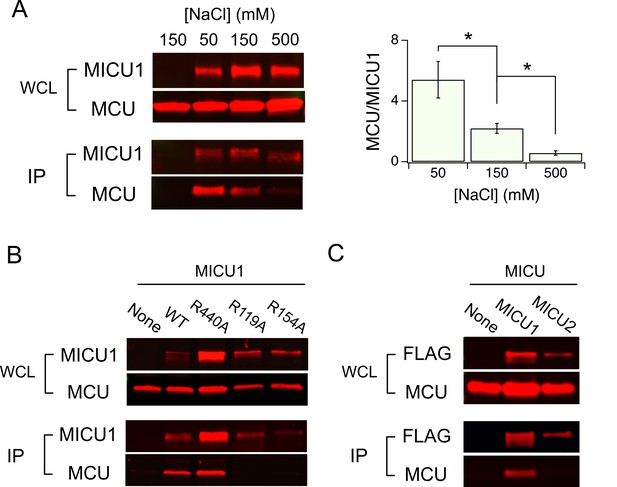
Electrostatic interactions between MCU and MICU1.
(A) Modulation of MCU-MICU1 complex stability by ionic strength. WT MCU and MICU1 were expressed in MCU/EMRE-KO cells, and CoIP experiments were performed in the presence of 50, 150, or 500 mM of NaCl. The IP signal of MCU was normalized to that of MICU1, with the ratio presented in the bar chart. (B) The effect of MICU1 Arg mutations on MCU binding. (C) A CoIP experiment testing if MCU and MICU2 form complexes. MICU2 was FLAG-tagged to precipitate WT MCU in MCU/EMRE-KO cells. *p<0.05.
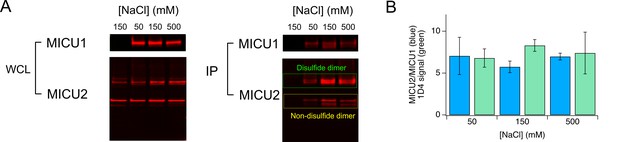
The effect of varying ionic strength on protein-protein interactions.
(A) CoIP experiments testing how increasing [NaCl] from 50 to 500 mM affects the stability of the non-disulfide MICU1-MICU2 complex. MICU1 is FLAG-tagged, and was used to pull down V5-tagged MICU2. The Western blot was performed in non-reducing conditions, showing that when MICU2 is overexpressed, it can form disulfide or non-disulfide heterodimers with MICU1 (Patron et al., 2014). The signal of MICU2 in the non-disulfide dimer is normalized to the signal of MICU1. (B) A data-summary bar chart. Data in (A) is presented as the ratio of MICU2 and MICU1 signals (Blue bars). Green bars represent the signal of 1D4-tagged MCU bound to the anti-1D4 antibody (images not shown; intensity readings were divided by 1000).
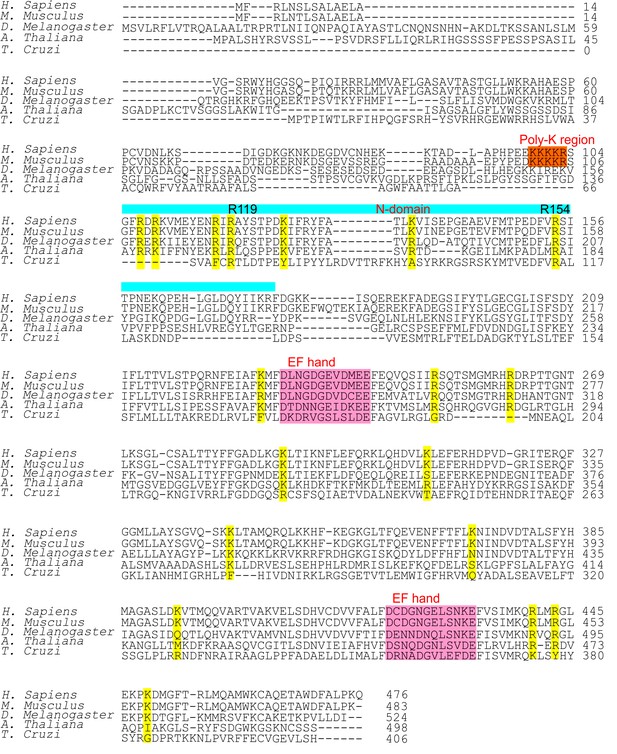
Multiple sequence alignment of MICU1.
Amino-acid sequences of ~120 MICU1 homologues in animals, plants, and protists were aligned. Positions that have either Arg or Lys in >70% of sequences were selected for a mutagenesis screen (yellow). Of these, only R119 and R154 (in human MICU1) are fully conserved. Two canonical EF hands, the N-terminal domain, and the poly-K EMRE binding region are also highlighted.
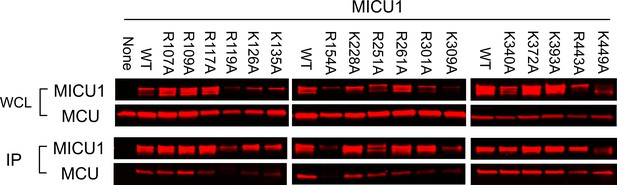
MICU1 mutagenesis screen.
FLAG-tagged, Arg or Lys mutants of MICU1 were expressed with 1D4-tagged WT MCU in MCU/EMRE-KO cells. CoIP shows that only R119A and R154A mutations abolish MCU binding. As these two mutants exhibit lower expression levels than WT, we further verified these results in Figure 5B using more DNA for transfection.
MICU1-MICU2 Interactions.
FLAG-tagged MICU1 constructs were co-expressed with V5-tagged WT MICU2 in MCU/EMRE-KO cells. Like WT MICU1, R119A and R154A MICU1 are able to complex with MICU2.
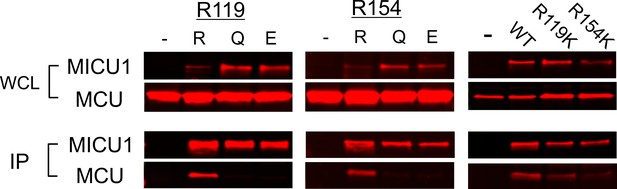
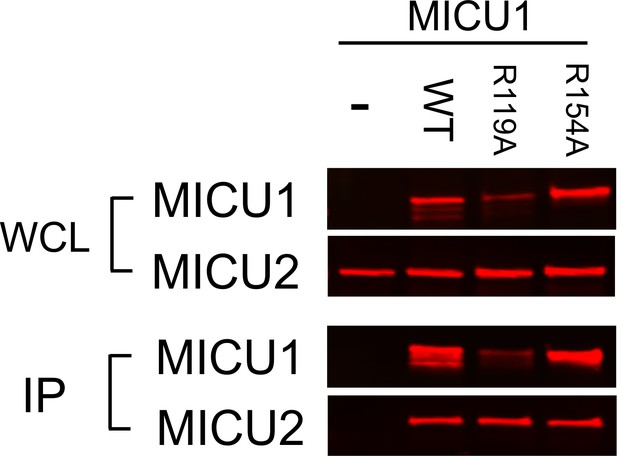
The effect of R119/R154 mutations on MCU-MICU1 complex formation.
Gln or Glu substitutions of R119 or R154 break the MCU-MICU1 complex, while R119K or R154K remain capable of forming a stable complex with MCU.
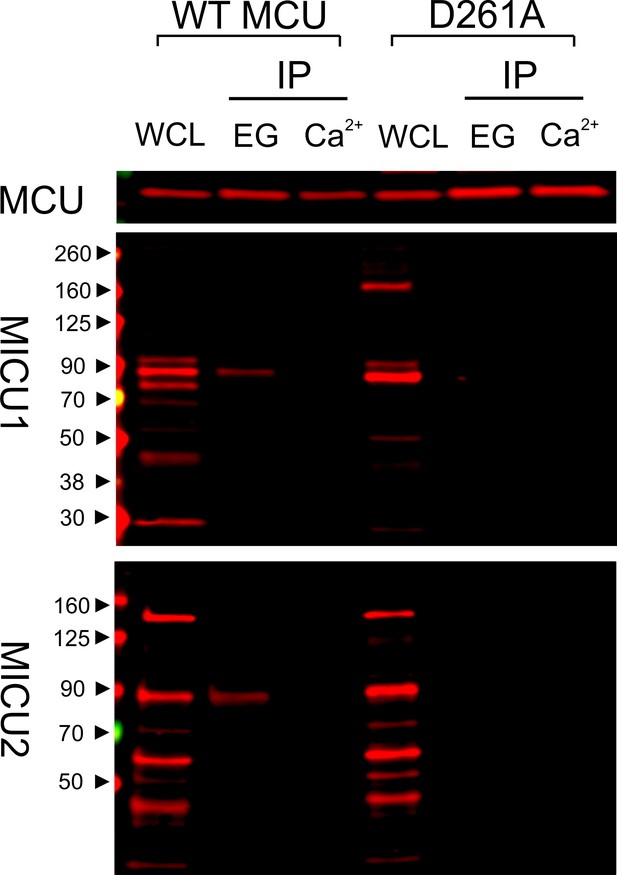
Ca2+-dependent interaction between MCU and the MICU1-2 heterodimer.
1D4-tagged WT or D261A MCU was expressed in WT HEK cells. The cell lysate, after a portion was taken for whole-cell lysate (WCL) analysis, was split into two for CoIP under Ca2+-free (EG, 1 mM EGTA) or 10 µM Ca2+ conditions. MCU was used to pull down the native, disulfide-connected MICU1-2 heterodimer (Patron et al., 2014; Petrungaro et al., 2015), which has a molecular weight of ~90 kDa. SDS-PAGE was performed in non-reducing environments. MICU1 and MICU2 were detected using anti-MICU1 and -MICU2 antibodies, respectively. WCL signals of MICU1 and MICU2 are not as clean as in previous images (e.g., Figure 2) due to the low abundance of native MICUs and lower qualities of these polyclonal MICU1 and MICU2 antibodies.
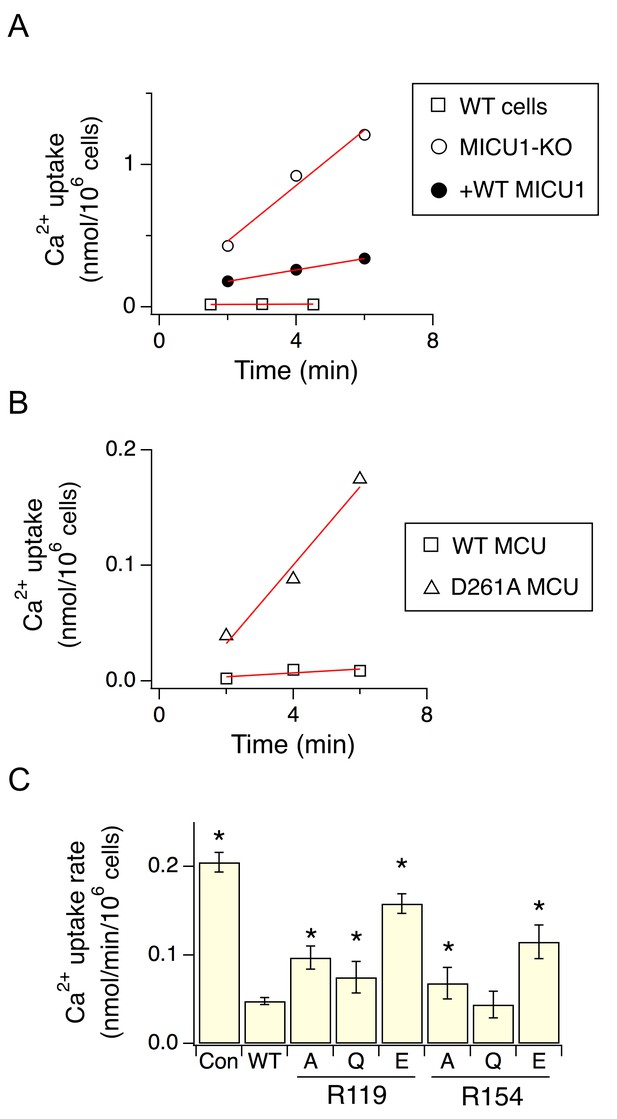
The effect of D261 or R119/R154 mutations on the regulatory function of MICU1.
(A) Mitochondrial Ca2+ uptake in a low Ca2+ (0.5 µM) condition. Each data point represents a measurement of 45Ca2+ transported into mitochondria by the uniporter at a specific time point. These data points were fit with a linear function (red lines) to obtain the rate of Ca2+ transport. (B) The activity of WT or D261A MCU in 0.5 µM Ca2+. (C) A bar chart summarizing the rate of mitochondrial Ca2+ uptake. WT MICU1 or various R119/R154 mutants were expressed in MICU1-KO cells. Con: untransfected control. Paired t-test was performed between WT MICU1 and mutants. *p<0.05.
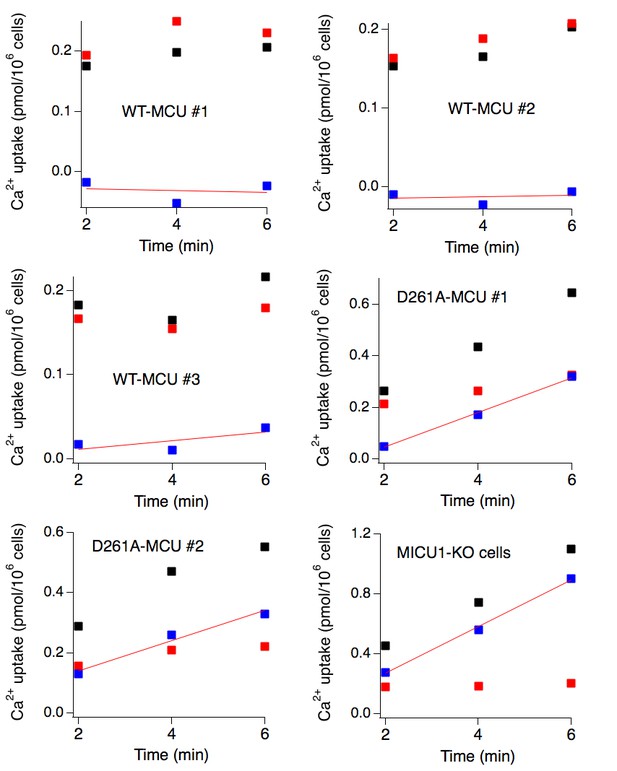
Data processing in 45Ca2+flux experiments.
6 independent 45Ca2+ flux (low Ca2+) experiments are presented. In each experiment, 45Ca2+ readings were obtained at three different time points (black squares). Non-specific signals (red squares) were obtained by adding Ru360, or by using untransfected cells for Ru360-insensitive mutants (e.g., D261A). Uniporter-specific signals (blue squares), obtained by subtracting non-specific signals (red squares) from total Ca2+ (black squares), were fit with a linear function (red lines) to produce the rate of Ca2+ transport. Rates from at least three experiments were then used for statistical analysis.
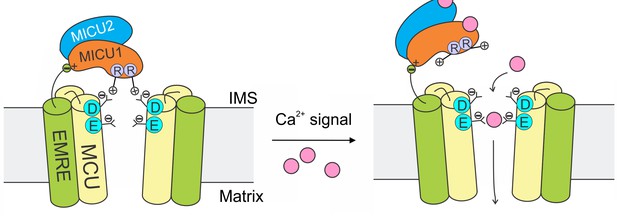
A model of Ca2+-dependent gating of the uniporter.
In resting cellular conditions, MICU1 shuts the uniporter by inserting Arg fingers into MCU’s Asp ring to occlude the pore. Ca2+ activates the channel by binding to MICUs to disrupt this MCU-MICU1 interaction. MICU2 forms a heterodimer with MICU1, but does not directly contact MCU. EMRE plays dual functional roles: it binds to MCU to enable Ca2+ permeation, and also interacts with MICU1 to maintain tight association of the MICU1-2 heterodimer with the uniporter during Ca2+ stimulation.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line | HEK 293T | ATCC | Cat # CRL-3216 | |
| Cell line | MCU-KO HEK 293T | PMID:27099988 | ||
| Cell line | MCU/EMRE-KO HEK 293T | PMID:27099988 | ||
| Cell line | MICU1-KO | PMID:28396416 | ||
| Primary Antibody | Mouse anti-FLAG | Sigma-Aldrich | Cat # F1804 | Western 1:10000 |
| Primary Antibody | Mouse anti-V5 | ThermoFisher | Cat # R960-25 | Western 1:5000 |
| Primary Antibody | Mouse anti-β actin | Santa Cruz | Cat # 69879 | Western 1:500 |
| Primary Antibody | Rabbit anti-MICU1 | Sigma-Aldrich | Cat # HPA037480 | Western 1:5000 |
| Primary Antibody | Rabbit anti-EFHA1 (MICU2) | Abcam | Cat # ab101465 | Western 1:10000 |
| Primary Antibody | Mouse anti-1D4 | PMID:6529569 | Western 50 ng/mL | |
| Primary Antibody | Mouse anti-C8 | PMID:8068416 | Western 50 ng/mL | |
| Secondary Antibody | IRDye 680RD goat anti-rabbit IgG | Li-Cor | Cat # 925–68073 | Western 1:10000 |
| Secondary Antibody | IRDye 680RD goat anti-mouse IgG | Li-Cor | Cat # 925–68072 | Western 1:15000 |
| Chemical compound | Ru360 | PMID:2036363 | ||
| Chemical compound | 45CaCl2 | PerkinElmer | Cat # NEX01300 | |
| Commercial kit | Lipofectamine 3000 | ThermoFisher | Cat # L3000015 | |
| Commercial kit | Anti-FLAG M2 affinity gel | Sigma-Aldrich | Cat # A2220 | |
| Commercial kit | CNBr-activated Sepharose 4B | GE Healthcare | Cat # 17043001 | |
| Software | Igor Pro 7 | WaveMetrics | Figure production and data fitting | |
| Software | ImageStudio 5 | Li-Cor | Western-blot quantification | |
| Software | Clustal Omega | PMID:21988835 | Sequence alignment | |
| Software | Excel (office 365) | Microsoft | t-test |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41112.021





