Structures in multiple conformations reveal distinct transition metal and proton pathways in an Nramp transporter
Figures
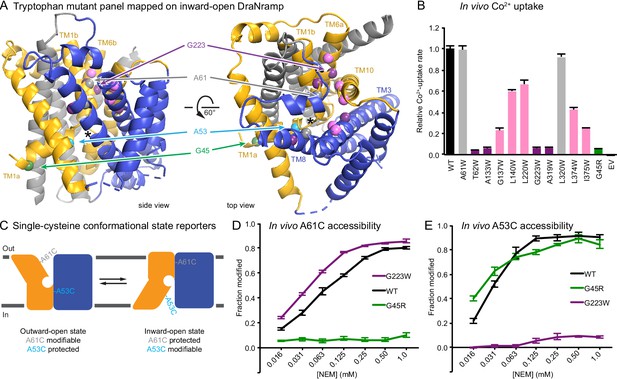
Design and validation of complementary conformationally-locked crystallization constructs.
(A) Panel of 11 designed tryptophan mutants (pink, purple, and gray spheres; see (B)) and G45R disease-mutant mimic (green) mapped onto our initial inward-open DraNramp structure and color-coded by their effect on transport activity in (B). Single-cysteine reporters A53C (cyan) and A61C (gray) are indicated. TMs 1, 5, 6, and 10 are colored gold, TMs 3, 4, 8, and 9 blue, and TMs 2, 7, and 11 gray. * marks the approximate location of the metal-binding site. (B) Relative in vivo Co2+ uptake rates for tryptophan mutant panel. Mutants colored gray, pink and purple did not affect, moderately decreased, or eliminated uptake, respectively. (C) Two complementary single-cysteine reporters enable assessment of the mutants’ conformational state preferences. For simplicity, in schematics DraNramp is depicted as two gold and blue domains that reorient to accomplish alternating access. (D) G45R prevented modification of the A61C outward-reporter, and (E) G223W fully protected the A53C inward-reporter, indicating that complementary conformational locking was achieved. Both reporters fully labeled for the WT-like protein, likely because it cycles between both conformations during the assay. Data in B, D, and E are averages ± S.E.M. (n = 4). See also Figure 1—figure supplement 1.
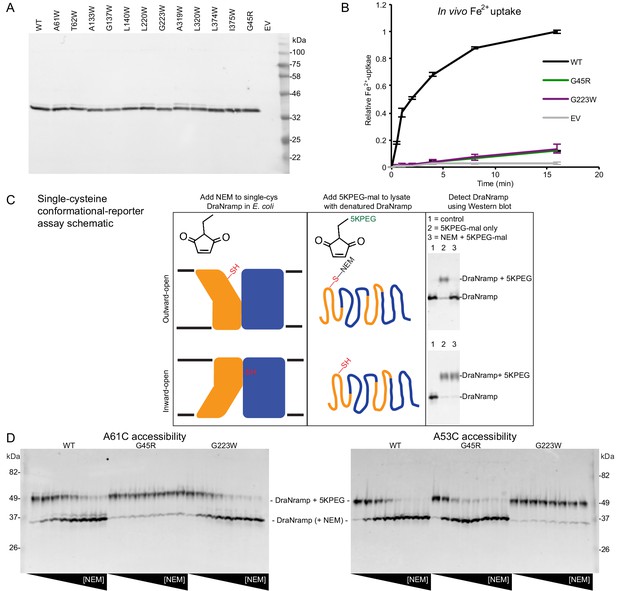
Validation of conformationally-locked crystallization constructs.
(A) Western blot against N-terminal His-tag for total lysates show most tryptophan mutants expressed similarly to WT DraNramp in E. coli. (B) Both G45R and G223W eliminated Fe2+ uptake in E. coli. (C) Schematic for cysteine accessibility assay two-step protocol using conformational reporters: first a gradient of NEM was applied to label any solvent-accessible cysteines, then the cells were lysed and protein denatured and 5K-PEG maleimide was added, which reacted with any previously-unmodified cysteines. The 5K-PEG maleimide-labeled protein shifted upward from the NEM-labeled protein on SDS-PAGE. (D) G45R and G223W had opposing effects on single-cysteine reporter accessibility to NEM modification in E. coli. The A61C reporter—only solvent-exposed in the outward-open state—was fully protected from NEM-labeling by the G45R mutation, indicating an inward-locked state. In contrast, A53C—likely only accessible in an inward-open conformation—was fully protected from NEM-labeling by G223W, indicating an outward-locked state. Both reporters were fully labeled at high NEM concentrations for WT, suggesting the entire protein population cycles through both conformational states during the NEM labeling time.
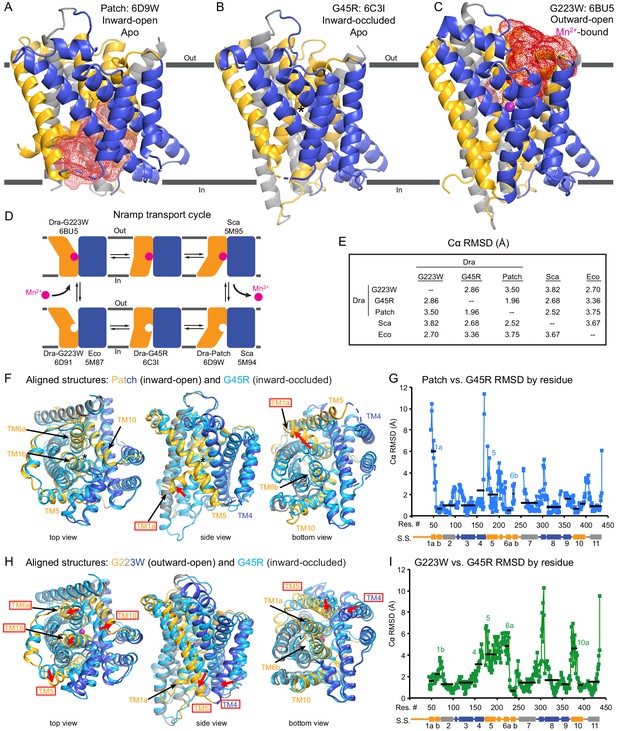
Crystal structures of DraNramp reveal two new conformations.
(A) Updated DraNramp Patch mutant structure in an inward-open apo state with a wide intracellular aqueous vestibule (red mesh) (Bozzi et al., 2016b). TMs 1, 5, 6, and 10 are colored gold, TMs 3, 4, 8, and 9 blue, and TMs 2, 7, and 11 gray. (B) The G45R structure revealed an apo inward-occluded conformation with no substantial extracellular or intracellular aqueous pathway to the metal-binding site, denoted by an * in structures where no metal is present. (C) The G223W structure revealed an outward-open conformation with the physiological substrate Mn2+ (magenta sphere) bound at the bottom of a substantial extracellular aqueous cavity (red mesh). All structures are viewed from within the membrane. (D) The complete Nramp transport cycle likely consists of at least six distinct conformational states. Including the structures of S. capitis (Sca) (Ehrnstorfer et al., 2014) and E. coleocola (Eco) (Ehrnstorfer et al., 2017) homologs, we now have structures of five of these conformations. (E) Pairwise RMSD values for the superposition of the 355 Cα atoms present in all three DraNramp structures, and ScaNramp and EcoNramp. G223W superimposes best with the outward-open EcoNramp, validating our mutagenesis strategy to obtain an outward-open conformation. G45R superimposes better with the inward-open Patch mutant than outward-open G223W, suggesting it represents an inward-occluded conformation. (F) Superposition of Patch mutant (gold, gray, and blue) and G45R (cyan) and (G) RMSD calculated at each Cα. These structures superimpose well except for TM1a. TM1a’s significant displacement from the inward-occluded state (G45R) to the inward-open state (Patch) is highlighted with a red arrow. (H) Superposition of G223W (gold, gray, and blue) and G45R (cyan) and (I) RMSD calculated at each Cα. The most significant rearrangements involve TMs 1b, 4, 5, 6a, and 10. These significant displacements from the inward-occluded state (G45R) to the outward-open state (G223W) are highlighted with red arrows. In panels G and I, black lines indicate average RMSD for each TM, with TMs 1, 6, and 10 divided into two halves. In panels F and H, the central view is rotated 45° along the vertical axis from the view in panels A-C, while the left and right views are 90° rotations of that central view. See also Figure 2—figure supplements 1 and 2.
-
Figure 2—source data 1
Nramp sequence alignment.
Alignment of 6878 Nramp sequences generated with HMMER and filtered to contain only sequences with canonical Nramp TM1 ‘DPGN’ and TM6 ‘MPH’ motifs.
- https://doi.org/10.7554/eLife.41124.009
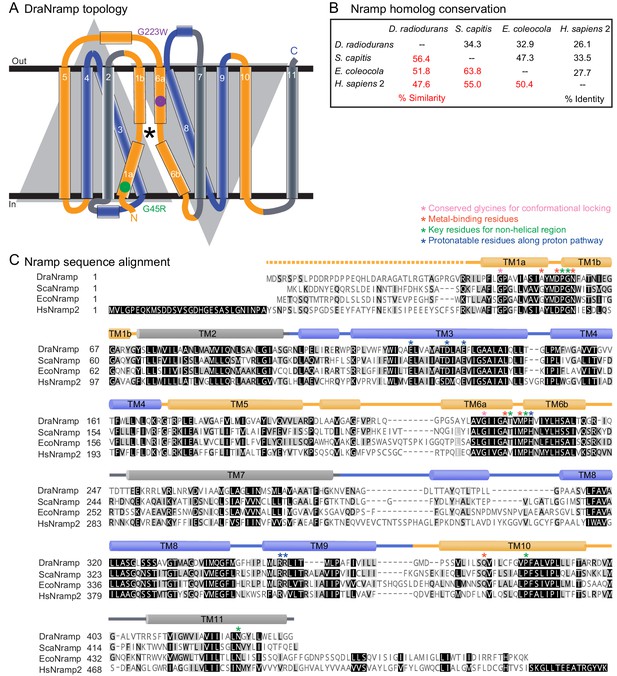
Secondary structure and primary sequence of DraNramp.
(A) DraNramp consists of 11 TMs, with the first ten TMs arranged in the canonical LeuT-fold: two pseudosymmetric inverted repeats of five TMs intertwine in the tertiary structure. The two conformational-locking mutations, G45R and G223W, on TMs 1a and 6a respectively, are at analogous positions within each repeat. The first and last TM in each repeat (1 and 5, and 6 and 10; gold) move significantly between the G223W and G45R structures, while the middle three remain relatively stationary (2–4, and 7–9; gray or blue). TMs 1, 4, 5, 6, and 10 group into a mobile domain, with the remaining six TMs considered a scaffold domain. * denotes the metal-binding site. (B) Sequence identity and similarity of DraNramp with S. capitis (Sca) and E. coleocola (Eco) Nramp homologs, which have been crystallized in the inward-open (Ehrnstorfer et al., 2014) and outward-open states (Ehrnstorfer et al., 2017) respectively, and human (Hs) Nramp2 (DMT1). Sequences were aligned using the BLOSUM62 matrix, with a cutoff of 1 used for the reported % similarity. (C) Sequence alignment of DraNramp, ScaNramp, EcoNramp, and HsNramp2 shows high conservation at positions of apparent structural significance and demonstrated functional importance.
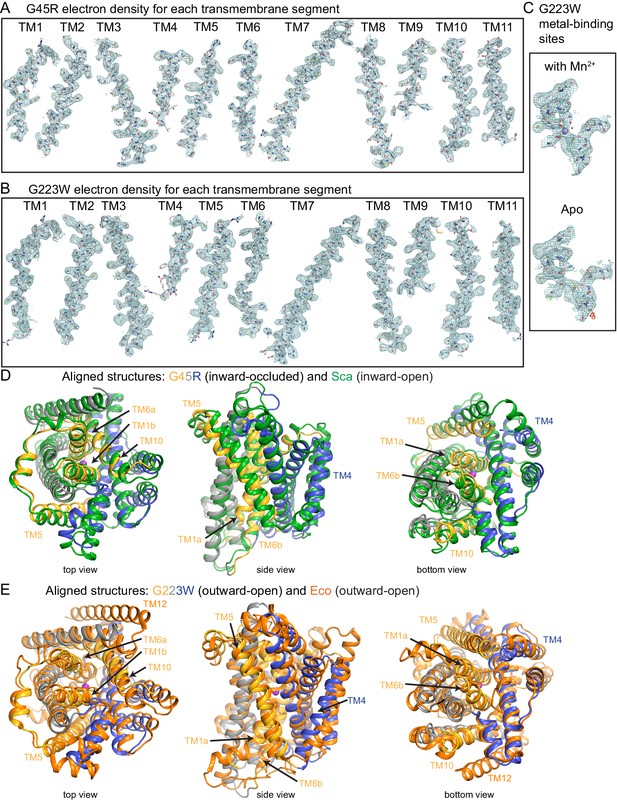
Electron density maps for new crystal structures and structural comparisons to other Nramp homologs.
2Fo-Fc electron density maps for each transmembrane segment of (A) G45R (6C3I) and (B) G223W (6BU5) structures contoured at 1.0 σ. (C) Electron density maps for the metal-binding site for the Mn2+-bound (6BU5) and apo (6D91) structures, with the latter also showing the Fo-Fc difference maps contoured at ±3.0 σ (green/red). Superpositions of (D) G45R with ScaNramp (green) and (E) G223W with EcoNramp (orange) confirm that our new structures represent outward-open and inward-occluded conformations, respectively. The views in (D) and (E) are based on a superposition of the G45R and G223W structures.
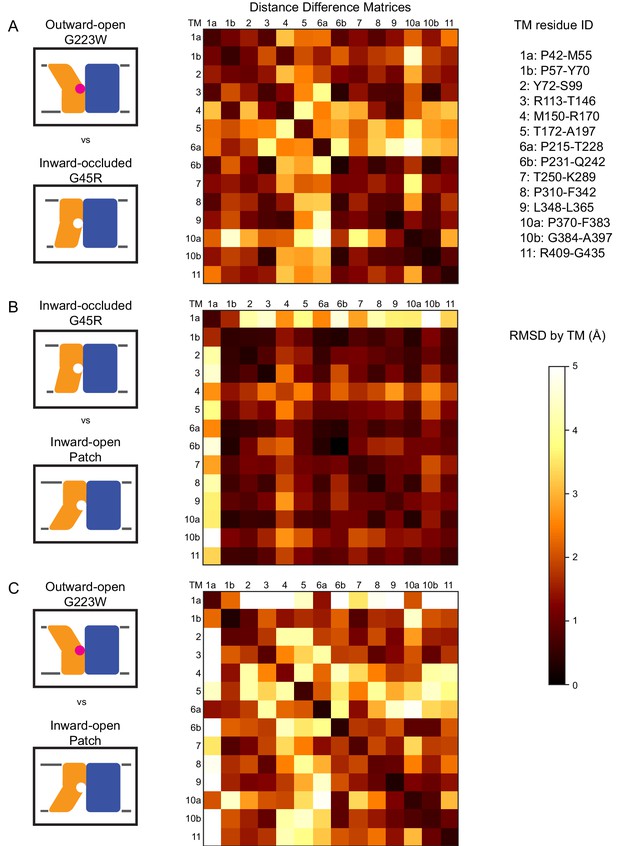
Distance difference matrices illustrate internal rearrangements during DraNramp conformational change.
We calculated the pairwise Cα – Cα distances to obtain a 398 × 398 distance matrix for each of the three DraNramp structures. To compare two structures, we generated distance difference matrices (Richards and Kundrot, 1988), subtracting one distance matrix from the other. To simplify the data and focus on relative movements between helices, we then calculated the root-mean-squared deviations of submatrices, grouping residues within each helical segment. We used a total of 14 segments (top right), breaking TMs 1, 6, and 10 into two separate segments before and after their helix-breaking elements. The resulting 14 × 14 matrices compare (A) outward-open G223W and inward-occluded G45R, (B) inward-occluded G45R and inward-open Patch mutant, and (C) outward-open G223W and inward-open Patch mutant. Pairs of helices that remain stationary relative to each other have RMSD values close to 0 indicated by darker colors. In contrast, pairs of helices that rearrange significantly relative to each other have larger magnitude values indicated by lighter coloring in the heat map. These matrices show that (A) TMs 4, 5, 6a, and 10a undergo the greatest displacement relative both to the rest of the protein and to each other in the conformational change from inward-occluded to outward-open. (B) In contrast, the conformational change from inward-open to inward-occluded consists primarily of the large displacement of TM1a, with the rest of the protein remaining mainly stationary. (C) The comparison of the outward-open and inward-open states is essentially a sum of the two previous comparisons, with the large TM1a displacement added to the significant movements of TMs 4, 5, 6a, and 10a. The Python code used to perform the calculations and generate the resulting plots is available at GitHub: https://github.com/GaudetLab/coarse-grained-DDMP.
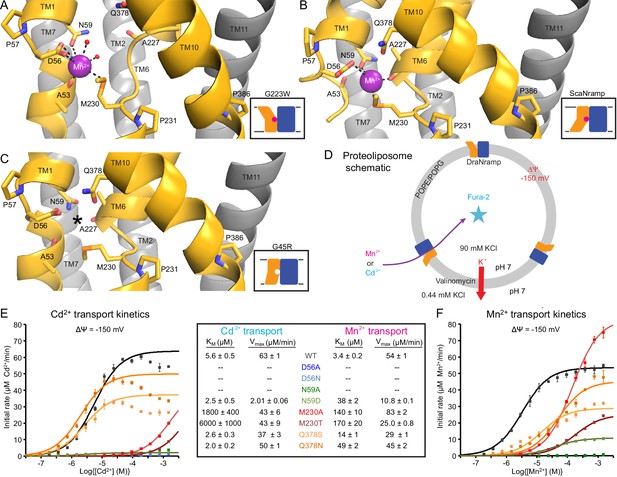
Metal-coordinating interactions vary across conformational states.
(A) In the G223W outward-open structure D56, N59, M230 and the A53 carbonyl, along with two water molecules, coordinate Mn2+. (B) In the inward-open ScaNramp structure (PDB: 5M95, DraNramp residue numbering) D56, N59, M230, and the A227 carbonyl coordinate Mn2+. (C) In the G45R inward-occluded structure D56, N59, M230, Q378, and the A53 and A227 carbonyls are all close, suggesting that all six could simultaneously coordinate the metal substrate in a hypothetical similar Mn2+-bound conformation. Three conserved helix-breaking prolines on TM1 (P57), TM6 (P231), and TM10 (P386) confer the flexibility needed for the metal-binding site. The TM6 unwound region is extended in the outward-open state, while TM10 bends more dramatically at P386 in the inward-oriented states to close outside access to the metal-binding site. For clarity, TMs 3, 4, 5, 8, and 9 are omitted in panels A-C. (D) Schematic for liposome transport assay to measure Mn2+ or Cd2+ uptake. (E–F) Plots of initial rates vs. (E) Cd2+ or (F) Mn2+ concentration used to determine Michaelis-Menten constants (Table inset) for WT DraNramp and binding-site mutants. All mutants significantly impaired Mn2+ transport at lower [Mn2+], while only Q378 mutants did not severely impair Cd2+ transport. Data are averages ± S.E.M. (n = 3); errors on KM and Vmax reflect uncertainty of the fit (shown as solid lines on the graphs) to the data. For WT and Q378 mutants we observed a reduced transport rate at high [Cd2+], which we did not include in the KM/Vmax fitting. These data are intriguing and remain unexplained, as we do not observe a similar effect for Mn2+, and we do not see an obvious second regulatory metal-binding site in our current structures of DraNramp. See also Figure 4—figure supplement 1.
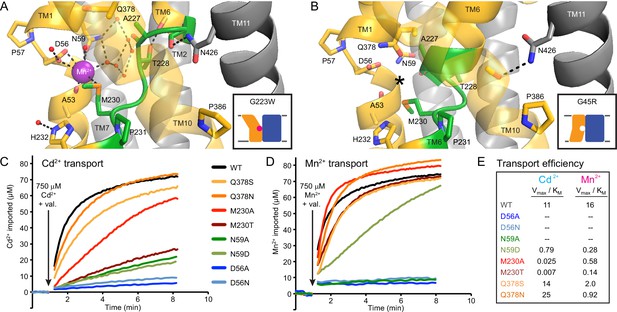
Extended TM6 unwound region enables metal-binding in outward-open DraNramp.
(A) TM6 unwinds in the outward-open G223W structure (non-helical region encompassing residues 226–231 is shown in green). Conserved sidechains on TM6 (T228) and TM11 (N426) donate hydrogen bonds to backbone carbonyls on TM6a to stabilize the extended non-helical region. A network of ordered water molecules forms the outer coordination sphere for the bound Mn2+ and extends into the outward metal-permeation pathway between TMs 1b, 6a, and 10. Conserved H232 below the metal-binding site also coordinates an ordered water. (B) For comparison the same region is highlighted in green in the G45R inward-occluded structure, although the TM6a helix is extended, and only residues 228–231 are unwound. Interestingly, in this structure the N426 sidechain donates a hydrogen bond to satisfy the backbone carbonyl on TM10 just above the conserved P386. For clarity, TMs 3, 4, 5, 8, and 9 are omitted and TM10 is shown as a transparent ribbon in panels A and B. (C–D) Representative traces (n = 3) for WT and binding-site mutants (C) Cd2+ and (D) Mn2+ uptake at 750 μM. Mutations to D56 eliminate all metal transport, while mutations to N59 severely impair transport for both metals. At higher metal concentrations, mutations to M230 are much more deleterious to Cd2+ transport than Mn2+ transport, while mutations to Q378 do not greatly impair transport of either metal. (E) Transport efficiency (Vmax/KM) comparisons from the data in Figure 4 illustrate all tested mutations are deleterious to Mn2+ transport, while all but Q378S and Q378N are deleterious to Cd2+ uptake.
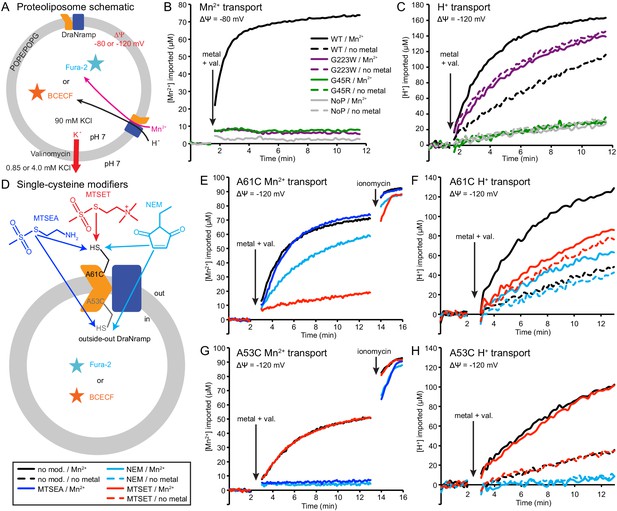
Outward-locked mutants retain H+ transport ability.
(A) Proteoliposome assay schematic for monitoring M2+ and H+ import. Membrane potential (ΔΨ) was established using K+ gradients and valinomycin. (B) Mn2+ uptake over time; WT enabled robust uptake while G45R and G223W showed no activity. NoP = No Protein control liposomes. (C) H+ uptake over time; WT enabled significant H+ uptake without metal (H+ uniport), further stimulated by Mn2+. G223W showed a larger H+ uniport rate, but Mn2+ provided no enhancement; G45R showed no H+ transport activity. Traces are representative of four experiments. (D) Schematic for in vitro cysteine modification of A53C (inside accessible) and A61C (outside accessible). MTSEA and NEM are membrane permeable, whereas permanently-charged MTSET is not. (E,G) Mn2+ and (F,H) H+ transport over time in the presence or absence of cysteine modifying agents. Adding ionomycin allows divalent cation entry to achieve maximum signal. Traces are representative of three experiments. See also Figure 5—figure supplements 1, 2 and 3.
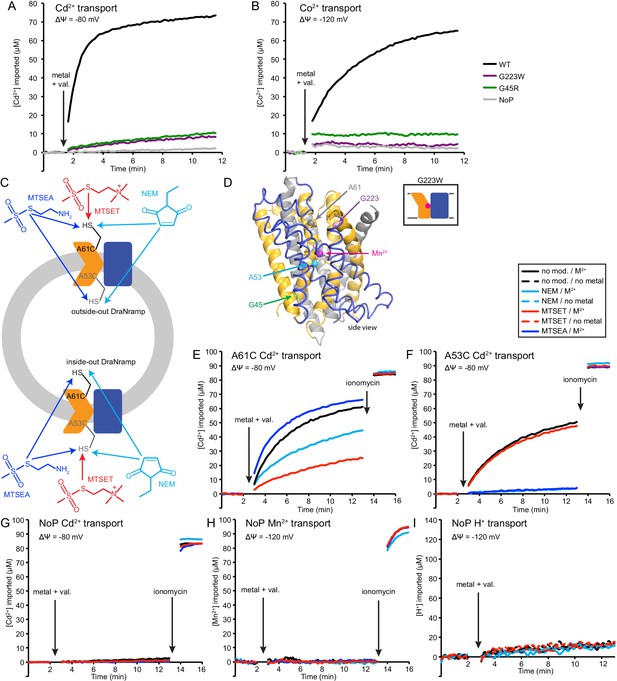
Conformational locking impairs metal transport.
Sample traces (n = 4) of (A) Cd2+ and (B) Co2+ transport over time in proteoliposome. WT enabled robust uptake while G45R and G223W did not. NoP = No Protein control liposomes. (C) Schematic for in vitro cysteine modification of A53C (inside accessible) and A61C (outside accessible). DraNramp randomly inserts into proteoliposomes, resulting in a ~ 50:50 mix of outside-out and inside-out transporters (Figure 5—figure supplement 2). A53C and A61C should thus be fully-labeled by membrane-permeable MTSEA and NEM for transporters in both orientations. In contrast, only outside-out A61C and inside-out A53C transporters should be labeled with the membrane-impermeable MTSET. (D) A61 and G223 line the metal permeation pathway to binding site, while A53 and G45 face the metal exit pathway to the cytoplasm. (E,F) Sample traces (n = 3) of Cd2+ transport over time in the presence or absence of cysteine modifying agents, with trends similar to Mn2+ transport in Figure 5E,G: (E) MTSET greatly reduced A61C Cd2+ transport, NEM had a moderate effect, and MTSEA had a slight enhancing effect; (F) NEM and MTSEA eliminated A53C Cd2+ transport, while MTSET had no effect. (G–I) Sample traces (n = 3) show that none of the modifiers enabled M2+ or H+ leak into empty liposomes.
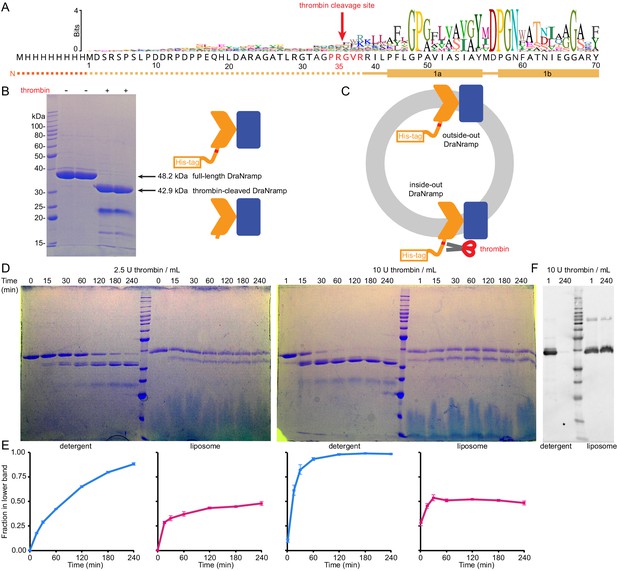
DraNramp inserts with a random orientation in proteoliposomes.
(A) Primary sequence of DraNramp from its N-terminus through TM1 along with sequence logo generated from our 6878-sequence alignment. DraNramp’s disordered N-terminus includes a predicted thrombin cleavage site: the ‘PRGVR’ sequence (red) that follows the P-R-[S/A/G/T]-[not D/E]-R pattern shown to provide optimal thrombin cleavage (Gallwitz et al., 2012). (B) Incubation of 0.9 mg/mL DraNramp in detergent with 17 units (U)/mL of human thrombin for 2 hr at room temperature leads to complete cleavage of the full-length protein, with the major product resulting from the removal of the first 35 native residues as well as the N-terminal His-tag. (C) DraNramp can hypothetically reconstitute in either an outside-out or inside-out orientation in proteoliposomes. The thrombin cleavage site is accessible to externally added thrombin for inside-out but not outside-out DraNramp. (D,E) Representative Coomassie-stained SDS-PAGE gels (D) and fraction of protein in the lower band (E) show the time course of 2.5 U/mL or 10 U/mL thrombin cleavage of 0.1 mg/mL detergent-solubilized and liposome-embedded DraNramp. Data are averages ± S.E.M. (n ≥ 3) corrected for the difference in expected molecular weights of the upper (48.2 kDa) and lower (42.9 kDa) bands. Thrombin cleavage of DraNramp reconstituted into proteoliposomes yields an equilibrium ~50:50 mix of full-length and cleaved protein, but fully cleaves DraNramp in detergent. (F) Western blot using a fluorescent anti-His antibody confirms complete removal of the N-terminus for DraNramp in detergent, while proteoliposome DraNramp retains significant full-length protein.
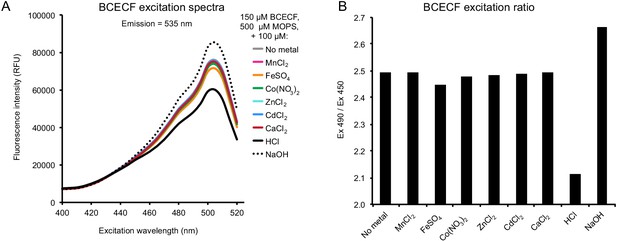
Divalent metals have little-to-no effect on BCECF fluorescence.
To mimic conditions inside of the liposomes, 150 μM BCECF in 0.5 mM MOPS, 90 mM KCl, 30 mM NaCl was mixed with 100 μM of a range of divalent cation salts as well as HCl and NaOH as controls. Only Fe2+ showed any significant perturbation of BCECF fluorescence, and with only a minor effect compared to adding the same concentration of H+ or OH-, as seen in (A) BCECF fluorescence spectra and (B) Ratio of fluorescence excitation at 490 nm and 450 nm. All data are averages of n = 4 technical replicates.
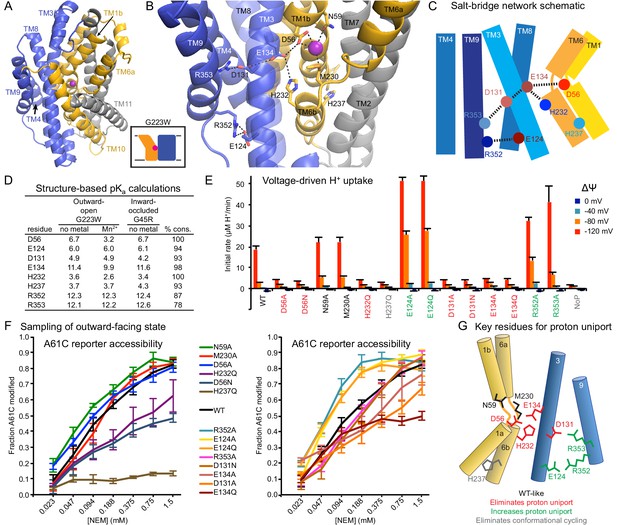
Conserved salt-bridge network provides pathway for proton uniport.
(A) View from an angle above the membrane looking down into the extracellular aqueous cavity in the outward-open G223W structure. (B) Zoomed-in view (with TM10 and TM11 omitted for clarity) and (C) schematic showing a network of conserved protonatable residues that originates from the metal-binding site and extends into the cleft between TMs 3, 4, 8, and 9. H232 and E134 abut metal-binding M230 and D56, providing a connection to the D131-R353 and R352-E124 salt-bridge pairs. (D) pKa estimates by PROPKA (Dolinsky et al., 2004) from high-resolution DraNramp structures. (E) Initial rates of basal H+ uptake at various ΔΨs (averages ± S.E.M.; n ≥ 4). Mutations to D56, E134, H232, and D131 eliminated H+ uniport observed for WT. N59 and M230 mutants retained significant H+ uptake. Mutations to E124, R352, and R353 enhanced H+ uniport. (F) Dose-response curve of outward-open reporter A61C modification versus NEM concentration. All mutants except H237Q sampled the outward-open state, which is required for H+ transport to occur (Figure 5). Data are averages ± S.E.M. (n ≥ 3). (G) Salt-bridge network schematic shows clustering of four residues required for H+ uniport (red) as well as three residues that may restrain this flux (green). See also Figure 6—figure supplement 1.
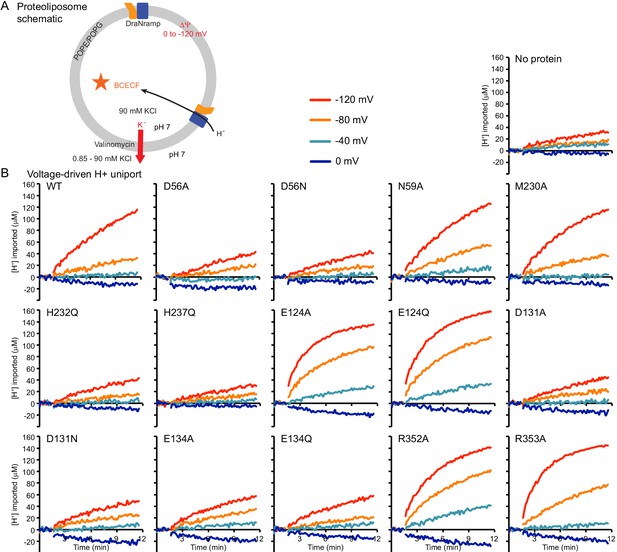
Sample traces illustrate mutant perturbations of voltage-driven proton uniport.
(A) Schematic for proteoliposome proton uptake assay. (B) Representative time traces (n ≥ 4) of H+ uptake into liposomes measured at four ΔΨ values for each mutant or no-protein control.
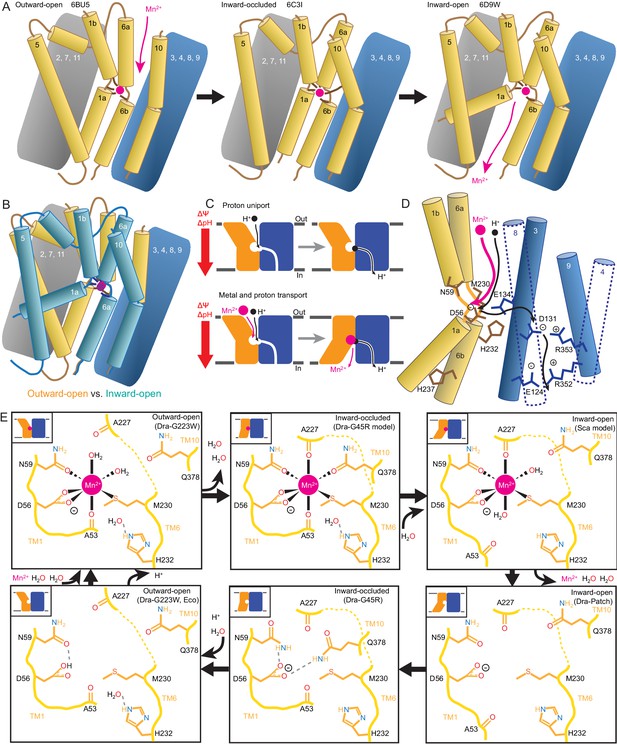
Mechanistic models for conformational change, proton transport, and metal transport.
(A) Model of conformational change: The metal ion reaches the outward-open binding site via the vestibule between TMs 1b, 6a, 10, 3, and 8. TM6a and 10 bend inward to close the vestibule in the inward-occluded conformation. TM6a’s inward bend pulls on TM5 via extracellular loop 5–6 to initiate opening of the inner gate. Finally, TM1a swings away from TM6b to open the intracellular vestibule into which the metal diffuses. (B) Superposition of outward- (gold) and inward-open (teal) conformations illustrate key movements by TMs 1, 5, 6, and 10. (C) Overall DraNramp transport model under prevailing physiological conditions (negative inside membrane potential, acidic-outside pH gradient): While metal transport requires complete conformational cycling, proton uniport occurs through the outward-open state. Metal-stimulated proton transport may follow the same pathway as proton uniport. (D) Proton transport model: A proton transits the external vestibule to reach the binding site near the membrane center and initially binds to D56. Metal enters through the same passageway, ejecting the proton, which then passes to D131, with H232 and E134 facilitating the transfer. The proton ultimately reaches the cytoplasm through the polar network between TMs 3, 4, 8, and 9, while the metal must await a bulk conformational change that opens a separate pathway between TMs 1a, 2, 5, 6b, 7, and 8. Proton uniport follows the same route, with the D56-D131 transfer occurring at a slower rate without metal substrate. (E) Model for metal coordination during the transport process: the metal initially binds in the outward-open state to D56, N59, M230 sidechains and the A53 carbonyl, shedding all but two water ligands. As the outward metal-permeation pathway closes, Q378 and the A227 carbonyl displace the waters so that the metal is fully coordinated by six amino acids in the inward-occluded state. As the inner gate opens, Q378 and the A53 carbonyl withdraw and are replaced by waters. The metal is then released into the cytoplasmic aqueous vestibule. To facilitate the return to the outward-open state, Q378 and N59 donate hydrogen bonds to negatively-charged D56 in the inward-occluded return conformation seen in our G45R structure. Finally, as the transporter returns to an outward-open state, it may bind a proton in preparation for metal-binding. For the inward-open metal-bound model for DraNramp, we altered the reported coordination from the Mn2+-bound ScaNramp relatively low-resolution (3.4 Å) crystal structure (PDB: 5M95) by adding two metal-coordinating waters, as well as switching the aspartate coordination to bidentate and the asparagine coordination to monodentate to be consistent with our G223W structure.
Videos
Internal rearrangements during DraNramp conformational changes.
Morph of the structure of DraNramp based on a global superimposition of the three captured conformational states. TMs 1, 5, 6, and 10 are colored gold, TMs 3, 4, 8, and 9 blue, and TMs 2, 7, and 11 gray, viewed in the membrane plane and the intracellular face pointing down. The morph starts from the G223W outward-facing conformation, transitions to the G45R occluded state, then to the Patch mutant inward-open state. The view then shifts to the intracellular face, followed by the external face, continuing to alternate back and forth through these three conformations.
Tables
DraNramp data collection and refinement statistics.
https://doi.org/10.7554/eLife.41124.005| Inward-open apo (Patch mutant + Fab) | Inward-occluded apo (G45R) | Outward-open Mn2+-bound (G223W ∆N34) | Outward-open apo (G223W ∆N34) | |
|---|---|---|---|---|
| PDB ID | 6D9W | 6C3I | 6BU5 | 6D91 |
| SBGrid Data Bank ID | 332, 333, 334 | 567 | 564 | 576 |
| Data Collection | ||||
| Wavelength (Å) | 1.139 | 0.9793 | 0.9792 | 0.9792 |
| Resolution range (Å) | 46.47–3.94 (4.08–3.94) | 50.00–3.00 (3.05–3.00) | 39.19–2.40 (2.49–2.40) | 38.14–2.36 (2.44–2.36) |
| Space group | I222 | P212121 | C121 | C121 |
| Unit cell (a, b, c) | 113.13, 132.08, 221.0 | 70.73, 97.85, 118.26 | 105.76, 80.39, 51.75 | 101.92, 75.51, 53.15 |
| Unit cell (α, β, γ) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 94.72, 90.0 | 90.0, 98.07, 90.0 |
| Number of crystals | 3 | 2 | ~15 | ~20 |
| Total reflections | 245867 | 66707 | 49321 | 48659 (367) |
| Unique reflections | 11791 (462) | 17206 (842) | 13983 (623) | 10998 (256) |
| Redundancy | 16.6 (11.4) | 3.9 (4.0) | 3.5 (1.7) | 4.4 (1.4) |
| Completeness (%) | 95 (96) | 98.5 (98.8) | 82.1* (36.4) | 64.0* (15.6) |
| Mean I/σ(I) | 6 (.61) | 6.41 (0.95) | 6.25 (1.56) | 8.83 (1.45) |
| Rmerge | 0.171 | 0.223 (1.388) | 0.186 | 0.185 |
| Rmeas | 0.177 | 0.270 (1.608) | 0.210 | 0.203 |
| Rpim | 0.047 | 0.133 (0.795) | 0.095 | 0.080 |
| CC1/2 | 0.99 (0.189) | 0.993 (0.396) | 0.984 (0.584) | 0.986 (0.683) |
| Refinement | ||||
| Resolution range (Å) | 46.47–3.94 (4.12–3.94) | 30.0–3.0 (3.05–3.00) | 39.19–2.40 (2.49–2.40) | 39.19–2.36 (2.44–2.36) |
| Rwork | 0.271 (0.253) | 0.244 (0.328) | 0.237 (0.269) | 0.244 (0.307) |
| Rfree | 0.309 (0.343) | 0.270 (0.394) | 0.276 (0.358) | 0.299 (0.437) |
| Number of atoms | 5764 | 5979 | 3219 | 3115 |
| Protein | 5761 | 5812 | 3012 | 3012 |
| Ligand | 3 | 138 | 143 | 86 |
| Water | -- | 29 | 64 | 17 |
| Protein residues | 786 | 774 | 398 | 398 |
| Ramachandran plot | ||||
| Favored (%) | 754 (96.0) | 762 (98.4) | 389 (97.7) | 381 (95.7) |
| Allowed (%) | 32 (4.0) | 12 (1.6) | 9 (2.3) | 17 (4.3) |
| Outliers (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| RMS(bonds) | 0.004 | 0.002 | 0.002 | 0.002 |
| RMS(angles) | 1.02 | 0.50 | 0.43 | 0.57 |
| Average B-factor | 90.3 | 61.27 | 73.6 | 50.1 |
| Protein | 90.3 | 61.29 | 73.1 | 49.9 |
| Ligand | 197.0 | 66.16 | 89.3 | 50.7 |
| Water | -- | 34.08 | 63.7 | 78.8 |
| Number of TLS groups | 15 | 20 | 6 | 7 |
-
*The G223WdN34 dataset compiled from many small crystals is 99% complete to 3.4 Å, 95% complete to 3.0 Å, 90% complete to 2.85 Å, 80% complete to 2.7 Å, and 60% complete to 2.5 Å. The G223W ΔN34 apo dataset compiled from many small crystals is 96% complete to 4.5 Å, 92% complete to 3.4 Å, 77% complete to 3.0 Å, 66% complete to 2.85 Å, and 52% complete to 2.7 Å.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers |
|---|---|---|---|
| Gene (Deinococcus radiodurans) | DraNramp | Genomic DNA | Uniprot: Q9RTP8 |
| Strain (Escherichia coli C41(DE3)) | C41(DE3) | Lucigen | 60442–1 |
| Recombinant DNA reagent | pET21a | Novagen | 69740–3 |
| Antibody | Alexa 647-conjugated anti-His-tag | Qiagen | 35370 |
| Chemical compound | DDM | Anatrace | D310S |
| Chemical compound | DM | Anatrace | D322S |
| Chemical compound | LMNG | Anatrace | NG310 |
| Chemical compound | Monoolein | Anatrace | LCP18 |
| Chemical compound | Fura-2 | Life Technologies | F-1200 |
| Chemical compound | BCECF | VWR | 102987–506 |
| Chemical compound | POPE | Avanti Polar Lipids | 850757C |
| Chemical compound | POPG | Avanti Polar Lipids | 840457C |
| Chemical compound | Valinomycin | Sigma-Aldrich | V0627 |
| Chemical compound | 5K-PEG maleimide | Creative PEGWORKS | PLS-234 |
| Chemical compound | NEM | Sigma-Aldrich | 4260 |
| Chemical compound | MTSET | VWR | 89139–738 |
| Chemical compound | MTSEA | VWR | 89139–740 |
| Software, algorithm | HKL2000 | PMID: 27799103 | RRID:SCR_015547 |
| Software, algorithm | XDS | PMID: 20124692 | RRID:SCR_015652 |
| Software, algorithm | PHASER | PMID: 19461840 | RRID:SCR_014219 |
| Software, algorithm | PHENIX | PMID: 22505256 | RRID:SCR_014224 |
| Software, algorithm | coot | PMID: 20383002 | RRID:SCR_014222 |
| Software, algorithm | PyMOL | RRID:SCR_000305 | |
| Software, algorithm | PROPKA | PMID: 15215472 | |
| Software, algorithm | HMMER | PMID: 21593126 | RRID:SCR_005305 |
| Software, algorithm | MUSCLE | PMID: 19461840 | RRID:SCR_011812 |
| Software, algorithm | ImageJ64 | RRID:SCR_003070 | |
| Software, algorithm | MATLAB | RRID:SCR_001622 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41124.021





