Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein
Figures
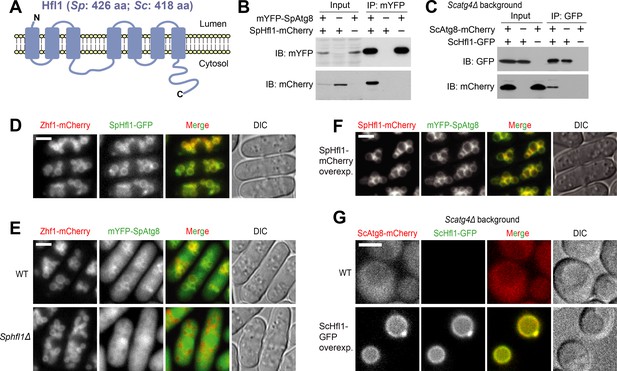
Hfl1 interacts with Atg8 and recruits Atg8 to the vacuole membrane.
(A) Schematic depicting the transmembrane topology of Hfl1 and its related proteins, as predicted using PolyPhobius (see Figure 1—figure supplement 1A for a sequence alignment). (B) SpHfl1 was co-immunoprecipitated with SpAtg8. (C) ScAtg8 was co-immunoprecipitated with ScHfl1. Scatg4Δ background was used to prevent the processing of ScAtg8-mCherry. (D) SpHfl1 localized to the vacuole membrane. Zhf1 is a vacuole membrane marker. (E) mYFP-SpAtg8 exhibited both cytosolic and vacuole membrane localizations in the wild-type cells, and the vacuole membrane localization was abolished in Sphfl1Δ cells. (F) Overexpression of SpHfl1 using the nmt1 promoter resulted in the concentration of mYFP-SpAtg8 on the vacuole membrane. (G) Overexpression of ScHfl1 using the TEF1 promoter resulted in the concentration of ScAtg8-mCherry on the vacuole membrane. Scatg4Δ background was used to prevent the processing of ScAtg8-mCherry. Bars, 3 µm.
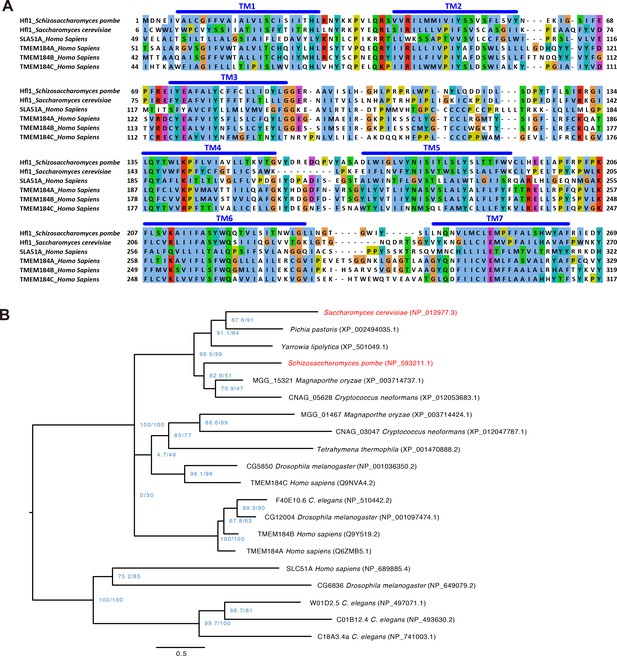
Sequence alignment and phylogenetic tree of Hfl1-related proteins.
(A) Multiple sequence alignment of the transmembrane regions of SpHfl1, ScHfl1, and four human proteins belonging to the InterPro IPR005178 protein family. Alignment was generated using MAFFT and visualized using Jalview. Transmembrane helices (TM) were predicted using PolyPhobius. (B) Maximum likelihood phylogenetic tree of IPR005178 protein family members in six fungal species (S. cerevisiae, P. pastoris, Y. lipolytica, S. pombe, M. oryzae, and C. neoformans), one ciliate species (T. thermophila), and three animal species (C. elegans, D. melanogaster, and H. sapiens). ScHfl1 and SpHfl1 are highlighted in red. Protein sequences were aligned using the L-INS-i iterative refinement algorithm of MAFFT and maximum likelihood analysis was performed using IQ-TREE version 1.5.3 for Mac OS X with the command line option -m TEST -alrt 1000 -bb 1000. The tree was rooted by midpoint rooting and visualized using FigTree. Blue-colored node labels are the SH-aLRT and the ultrafast bootstrap support values (in percentage) calculated by IQ-TREE. Scale bar indicates 0.5 substitutions per site.
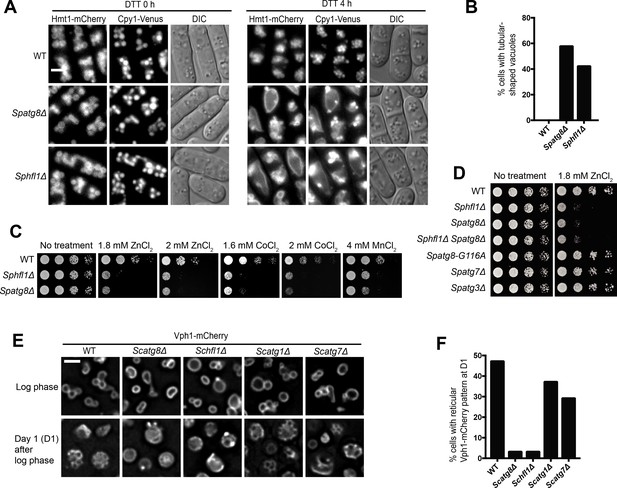
Hfl1 is required for the lipidation-independent vacuolar functions of Atg8.
(A, B) Micrographs (A) and quantitation (B) showing that DTT treatment induced the formation of tubular-shaped vacuoles in Spatg8Δ and Sphfl1Δ cells, but not in wild-type cells. Hmt1-mCherry and Cpy1-Venus are a vacuole membrane marker and a vacuole lumen marker, respectively. A representative result of three independent experiments is shown. (C) Spatg8Δ and Sphfl1Δ exhibited the same metal hyper-sensitivity. Five-fold serial dilutions of cells were spotted on a YES plate and YES plates with metal salts at the indicated concentrations. (D) Spatg8Δ Sphfl1Δ double mutant exhibited no enhanced metal sensitivity compared with the two single mutants, and mutants defective in Atg8 lipidation did not show metal hyper-sensitivity. See Figure 2—figure supplement 2 for the results of CoCl2 and MnCl2 treatment. (E, F) Micrographs (E) and quantitation (F) showing that Scatg8Δ and Schfl1Δ, but not Scatg1Δ and Scatg7Δ, were defective in stationary-phase-induced vacuolar microdomain formation. A representative result of three independent experiments is shown. Bars, 3 µm.
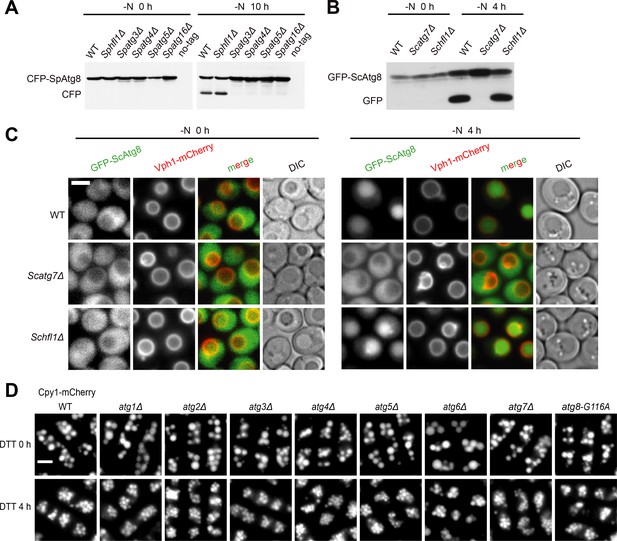
Hfl1 is not required for starvation-induced autophagy and fission yeast autophagy mutants other than Spatg8Δ do not exhibit vacuole morphology defect after DTT treatment.
(A) CFP-SpAtg8 processing assay showed that SpHfl1 is not required for starvation-induced autophagy. (B) GFP-ScAtg8 processing assay showed that ScHfl1 is not required for starvation-induced autophagy. (C) Imaging the localization of GFP-ScAtg8 showed that ScHfl1 is not required for starvation-induced autophagy. (D) Fission yeast autophagy mutants other than Spatg8Δ exhibited normal vacuole morphology upon DTT treatment. Bars, 3 µm.
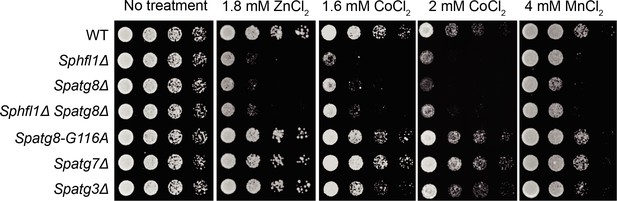
Spatg8Δ Sphfl1Δ double mutant exhibited no enhanced metal sensitivity compared with the two single mutants, and mutants defective in Atg8 lipidation did not show metal hyper-sensitivity.
https://doi.org/10.7554/eLife.41237.006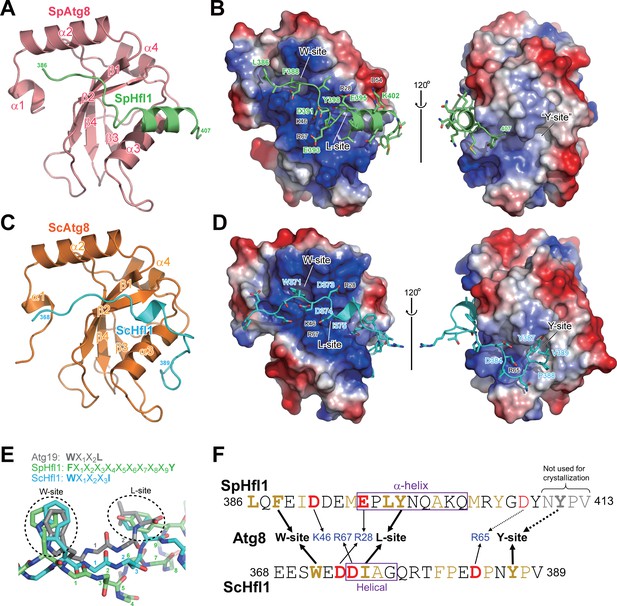
Atg8-Hfl1 binding is mediated by noncanonical AIMs and involves a previously unknown binding site on ScAtg8.
(A, C) Overall structures of the SpAtg8-SpHfl1(386-409) complex (A) and the ScAtg8-ScHfl1(368-389) complex (C) in ribbon diagrams. (B, D) Electrostatic surface potentials calculated for the surfaces of SpAtg8 (B) and ScAtg8 (D). (E) Structural comparison of a canonical AIM and the helical AIMs in SpHfl1 and ScHfl1. Crystal structures of ScAtg8-ScAtg19 (PDB 2ZPN; a representative structure of canonical AIMs bound to Atg8), SpAtg8-SpHfl1 and ScAtg8-ScHfl1 complexes were superimposed with each other by minimizing the rms differences of main-chain atoms of Atg8. The main chains of ScAtg19, SpHfl1, and ScHfl1 are shown with stick models, and the side chains of the two hydrophobic residues that bind to the W-site and the L-site are also shown. The residues intervening the two hydrophobic residues are numbered. (F) Summary of the interactions observed between Atg8 and Hfl1. The residues forming hydrophobic interactions are colored yellow, while those forming electrostatic interactions are colored blue (basic) and red (acidic). Hfl1 residues that when mutated affect the affinity with Atg8 by ITC measurements are highlighted with bold letters.
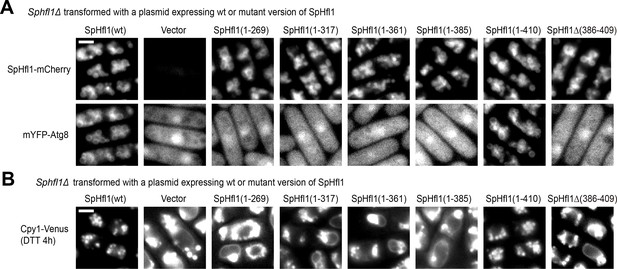
The abilities of SpHfl1 to recruit SpAtg8 and complement the vacuole morphology defect of Sphfl1Δ require its amino acids 386–409.
(A) Amino acids 386–409 of SpHfl1 are necessary for its ability to recruit SpAtg8 to the vacuole membrane. (B) Amino acids 386–409 of SpHfl1 are necessary for its ability to rescue the vacuole morphology defect of Sphfl1Δ. Bars, 3 µm.
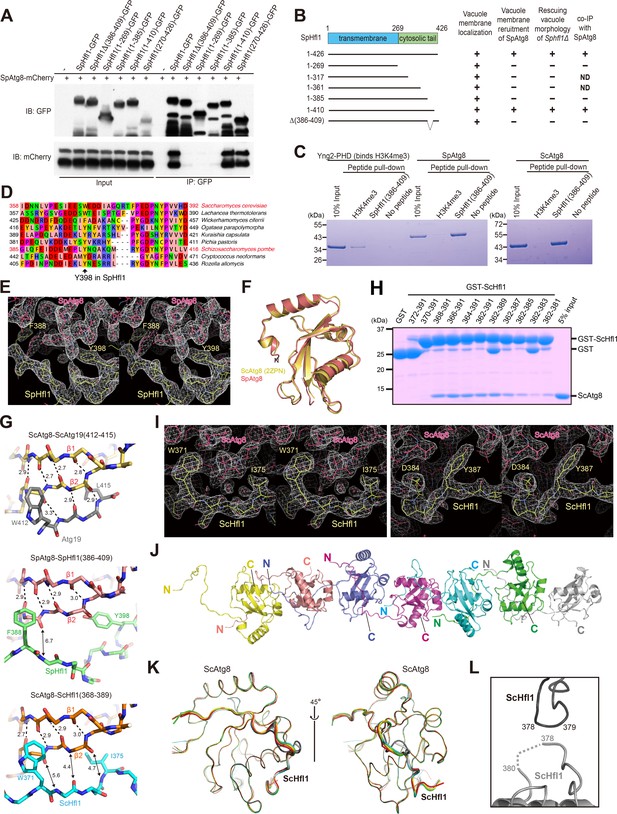
Mapping and structural analysis of the Atg8-binding regions in SpHfl1 and ScHfl1.
(A) Amino acids 386–409 in SpHfl1 are required for SpAtg8 binding. (B) Schematic summarizing the results shown in Figure 3—figure supplement 1A and B, and Figure 3—figure supplement 2A. ND, not determined. (C) Amino acids 386–409 in SpHfl1 are sufficient for specific in-vitro interactions with SpAtg8 and ScAtg8. (D) Sequences corresponding to the Atg8-interacting region of SpHfl1 in SpHfl1-related proteins from nine representative fungal species. SpHfl1-related proteins were identified using NCBI BLASTP and aligned using MAFFT. The alignment was manually adjusted and visualized using Jalview. The accession numbers of the sequences (from top to bottom) are: NP_012977.3, XP_002554458.1, XP_011275000.1, XP_013933679.1, CDK27935.1, XP_002494035.1, NP_593211.1, XP_012053683.1, and EPZ35735.1. (E, I) Stereo views of the 2Fo-Fc electron density maps for the SpAtg8-SpHfl1(386-409) complex (E) and the ScAtg8-ScHfl1(368-389) complex (I) calculated using COOT. (F) Structural comparison between SpAtg8 and ScAtg8. Crystal structure of ScAtg8 (PDB 2ZPN) was superimposed on that of SpAtg8 by minimizing the rms differences of main-chain atoms. (G) Intermolecular β-sheet is not formed between Atg8 and Hfl1. Crystal structures of the ScAtg8-ScAtg19(412-415) complex (top; PDB 2ZPN), the SpAtg8-SpHfl1(386-409) complex (middle), and the ScAtg8-ScHfl1(368-389) complex (bottom) are shown with stick models. Broken lines indicate hydrogen bonds responsible for forming the intra- and inter-molecular β-sheets. Numbers indicate distances in Å. For clarity, only main chains are shown except for the two hydrophobic residues that bind to the W-site and the L-site. (H) Amino acids 368–389 in ScHfl1 are important for strong in-vitro interactions with ScAtg8. (J) Head-to-tail interactions of seven copies of the ScHfl1(368-389)-ScAtg8 fusion protein observed in the asymmetric unit. (K) Superimposition of seven copies of the ScAtg8-ScHfl1 complex shown in a main-chain trace. Two copies, colored dark and light gray, show a slightly different conformation. (L) Crystal-packing interactions observed between two copies of ScHfl1. Dark and light gray copies correspond to those in (K).
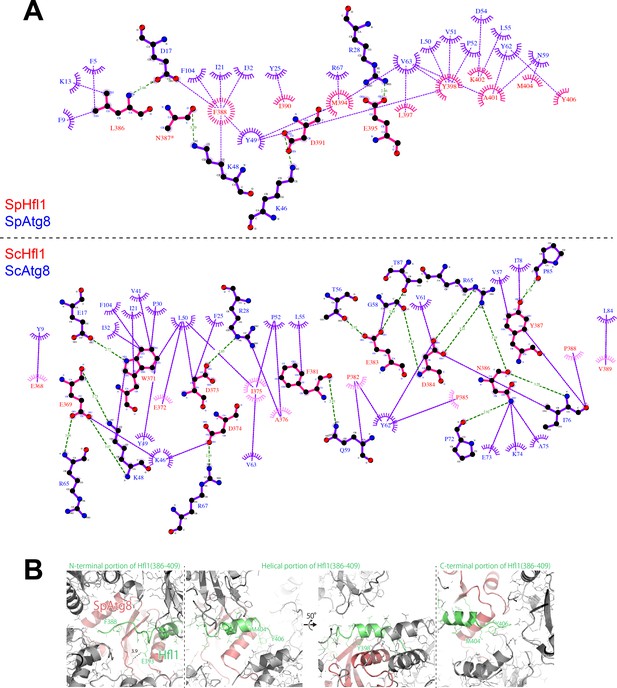
LigPlot+ diagrams and crystal packing of SpAtg8-SpHfl1(386-409).
(A) 2D protein interaction diagrams (LigPlot+ diagrams) of SpHfl1-SpAtg8 and ScHfl1-ScAtg8 complexes. (B) Crystal packing of SpAtg8-SpHfl1(386-409).
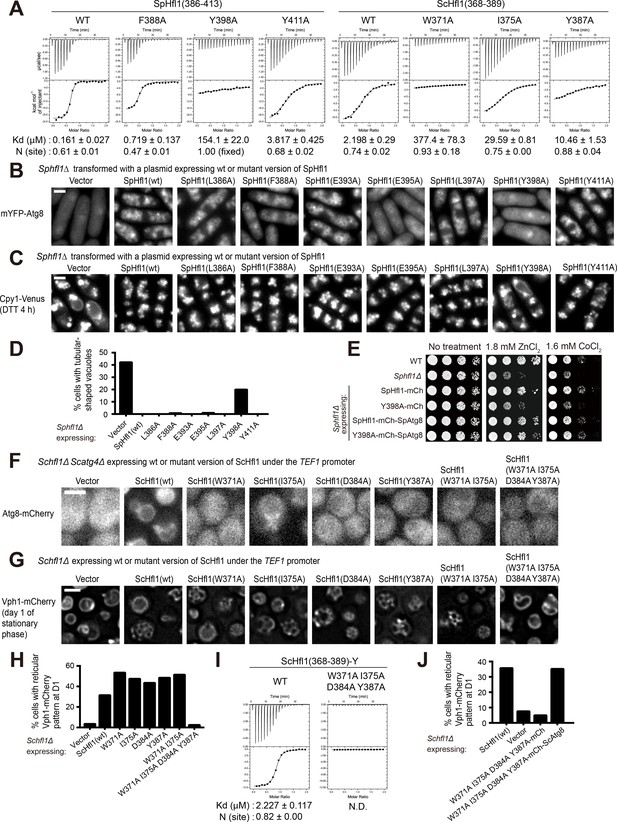
The Atg8-Hfl1 interaction is important for the lipidation-independent vacuolar functions of Atg8.
(A) ITC results obtained by titration of SpHfl1(386-413) into SpAtg8 or ScHfl1(368-389) into ScAtg8. (B) The effects of point mutations in the SpAtg8-binding region of SpHfl1 on the ability of SpHfl1 to recruit SpAtg8 to the vacuole membrane. (C) The effects of point mutations in the SpAtg8-binding region of SpHfl1 on the ability of SpHfl1 to complement the vacuole morphology phenotype of Sphfl1Δ. (D) Quantitation of the vacuole morphology phenotype shown in (C). A representative result of three independent experiments is shown. (E) Y398A mutation strongly diminished the ability of SpHfl1 to complement the metal hyper-sensitivity phenotype of Sphfl1Δ and fusion with SpAtg8 restored this ability. (F) The effects of point mutations in the ScAtg8-binding region of ScHfl1 on the ability of ScHfl1 to recruit ScAtg8 to the vacuole membrane. ScHfl1 was expressed using the strong TEF1 promoter. (G) The effects of point mutations in the ScAtg8-binding region of ScHfl1 on the ability of ScHfl1 to complement the microdomain formation phenotype of Schfl1Δ. ScHfl1 was expressed using the strong TEF1 promoter. (H) Quantitation of the microdomain formation phenotype shown in (G). A representative result of three independent experiments is shown. (I) ITC results obtained by titration of ScHfl1(368-389) with an additional Tyr residue at the C-terminus into ScAtg8. Addition of Tyr allowed the quantification of the concentration of the ScHfl1(W371A I375A D384A Y387A) by absorbance at 280 nm. (J) Fusing ScAtg8 to ScHfl1(W371A I375A D384A Y387A) restored the ability to complement the microdomain formation phenotype of Schfl1Δ. A representative result of three independent experiments is shown. Bars, 3 µm.
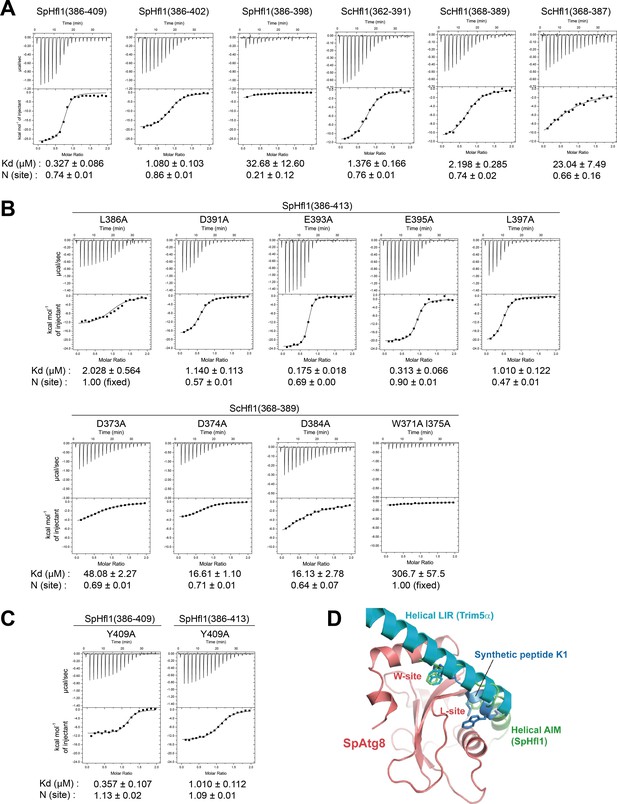
ITC experiments for identifying Hfl1 residues important for its binding to Atg8 and structural comparison with two other Atg8 binding sequences that possess a helical conformation.
(A) ITC results obtained by titration of SpHfl1 and ScHfl1 fragments of different lengths into SpAtg8 and ScAtg8, respectively. (B) ITC results obtained by titration of SpHfl1(386-413) and ScHfl1(368-389) mutants into SpAtg8 and ScAtg8, respectively. (C) ITC results obtained by titration of the Y409A mutant forms of SpHfl1(386-409) and SpHfl1(386-413) into SpAtg8, respectively. (D) Structural comparison with helical LIR and synthetic peptide K1. Crystal structures of LC3B-helical LIR complex (PDB ID 5W9A) and GABARAP-K1 complex (PDB ID 3D32) were superimposed on that of SpAtg8-SpHfl1 complex by minimizing the rms of main-chain atoms of Atg8-family proteins. LC3B and GABARAP were removed from the figure for clarity. The side-chains of hydrophobic residues that bind to the W-site and the L-site are shown with stick models.
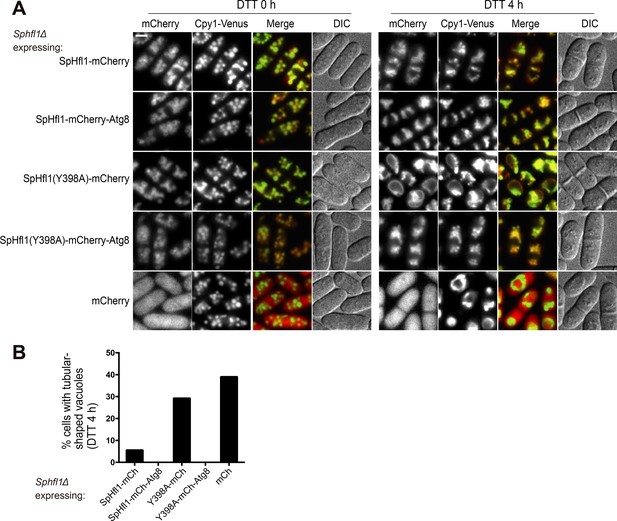
Fusion with SpAtg8 rescued the ability of SpHfl1-Y398A to complement the vacuole morphology defect of Sphfl1Δ.
(A) Assessing whether fusing with SpAtg8 can rescue the ability of SpHfl1-Y398A to complement the vacuole morphology defect of Sphfl1Δ. Bar, 3 µm. (B) Quantitation of the DTT 4 h data shown in (A). A representative result of three independent experiments is shown.
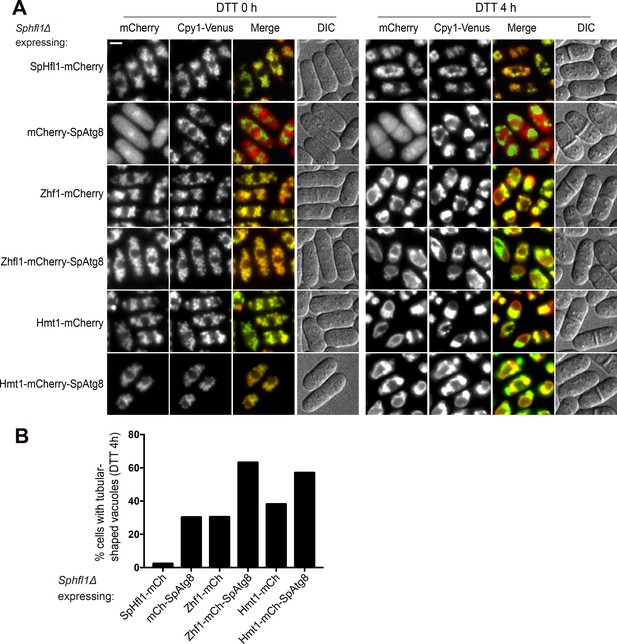
Fusing SpAtg8 with non-SpHfl1 vacuole membrane proteins did not bypass the requirement of SpHfl1.
(A) Assessing whether fusing SpAtg8 with non-SpHfl1 vacuole membrane proteins can rescue the vacuole morphology defect of Sphfl1Δ. Bar, 3 µm. (B) Quantitation of the DTT 4 h data shown in (A). A representative result of three independent experiments is shown.
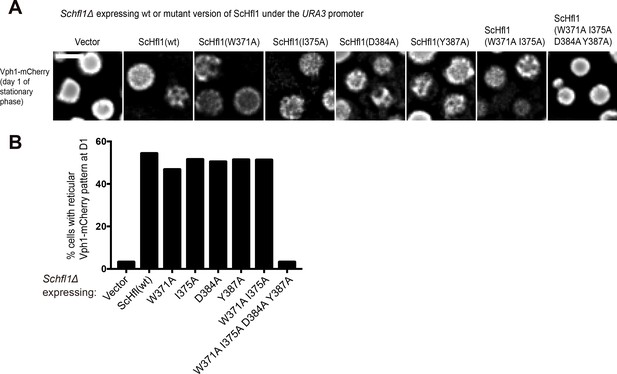
Assessing the ability of ScHfl1 mutants expressed from a weak promoter to complement the microdomain formation defect of Schfl1Δ.
(A) The effects of point mutations in the ScAtg8-binding region of ScHfl1 on the ability of ScHfl1 to complement the microdomain formation phenotype of Schfl1Δ. ScHfl1 was expressed using the weak URA3 promoter. Bar, 3 µm. (B) Quantitation of the microdomain formation phenotype shown in (A). A representative result of three independent experiments is shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Schizosaccharomyces pombe) | hfl1 | NA | PomBase: SPAC30D11.06c | |
| Gene (Saccharomyces cerevisiae) | HFL1 | NA | SGD:YKR051W | |
| Genetic reagent (Schizosaccharomyces pombe) | Fission yeast strains used in this study | this paper | See Supplementary file 2—Table S2 | |
| Genetic reagent (Saccharomyces cerevisiae) | Budding yeast strains used in this study | this paper | See Supplementary file 2—Table S3 | |
| Antibody | anti-GFP (mouse monoclonal) | Roche | Cat# 11814460001; RRID:AB_390913 | |
| Antibody | anti-mCherry (mouse monoclonal) | Abmart | ||
| Recombinant DNA reagent | Plasmids used for this study | this paper | See Supplementary file 2—Table S4 | |
| Commercial assay or kit | GFP-Trap agarose beads | ChromoTek | Cat# gta-20; RRID: AB_2631357 | |
| Commercial assay or kit | Pierce High Capacity Streptavidin Agarose | Thermo Fisher Scientific | Cat# 20359 |
Additional files
-
Supplementary file 1
Data of the affinity purification coupled with mass spectrometry (AP-MS) analysis.
- https://doi.org/10.7554/eLife.41237.016
-
Supplementary file 2
Supplementary tables listing the crystallographic data collection and refinement statistics, and the yeast strains and plasmids used in this study.
- https://doi.org/10.7554/eLife.41237.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41237.018





