Predicted glycosyltransferases promote development and prevent spurious cell clumping in the choanoflagellate S. rosetta
Figures
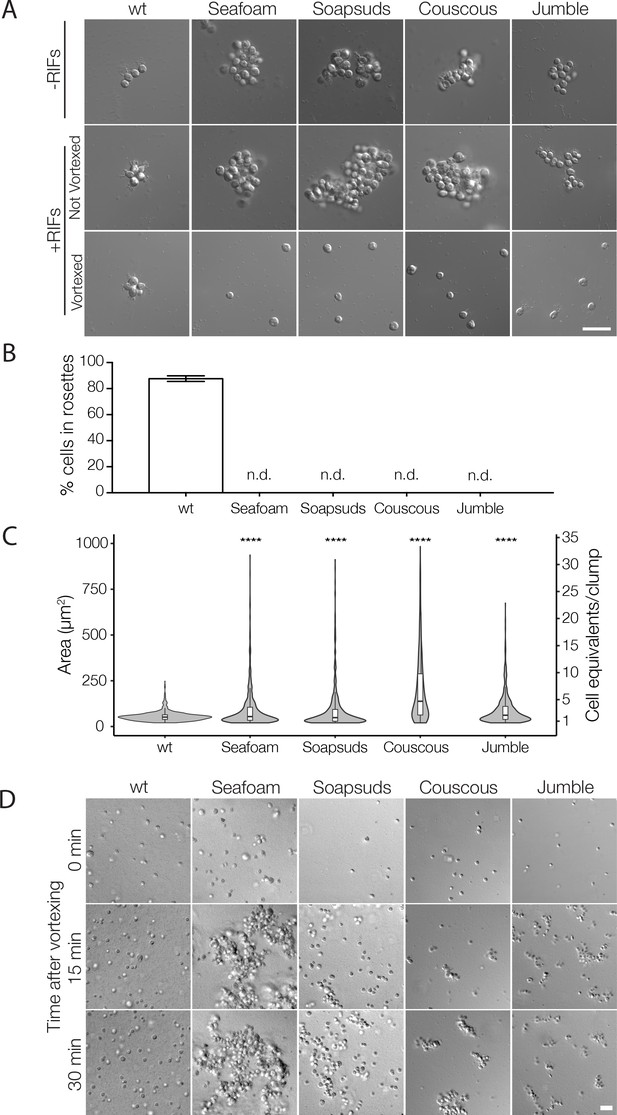
Mutant cells aggregate and fail to form rosettes.
(A) Wild type cells are unicellular or form linear chains in the absence of rosette inducing factors (RIFs) and develop into organized spherical rosettes when cultured with RIFs. Rosettes are resistant to shear force and survive vortexing. Four class C mutants — Seafoam, Soapsuds, Couscous, and Jumble — form disordered clumps of cells in the presence and absence of RIFs. The clumps are not resistant to vortexing and fall apart into single cells. (B) Class C mutants do not form any detectable rosettes. Rosette development was measured as the % of cells in rosettes after 48 hr in the presence of RIFs and is shown as mean ± SEM. n.d. = no detected rosettes. (C) Class C mutants quickly aggregated into large clumps after disruption by vortexing. After vortexing, wild type and mutant cells were incubated for 30 min in the absence of RIFs and clump sizes were quantified by automated image analysis. Data are presented as violin boxplots, showing the median cell number (horizontal line), interquartile range (white box), and range excluding outliers (vertical line). All mutants had significantly larger masses of cells (K-S test, ****p < 0.0001) than found in cultures of wild type cells. (D) Clumping occurred within minutes after vortexing in the Class C mutants without RIFs, revealing that the clumps form by aggregation and not through cell division. DIC images obtained at 0, 15, and 30 min post-vortexing. Scale bars = 20 μm.
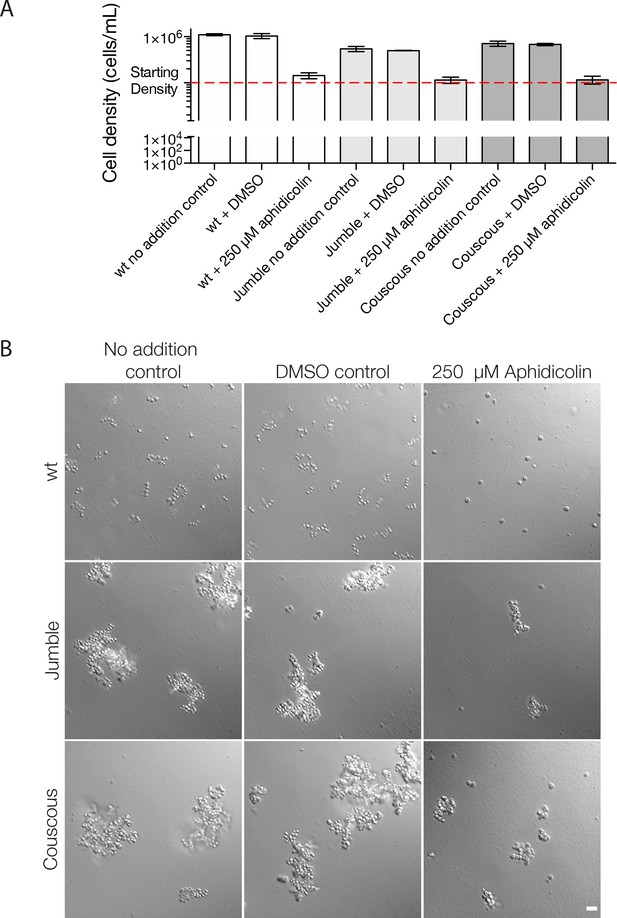
Cell division is not required for clump formation in mutants.
Wild type, Jumble, and Couscous cells were vortexed and diluted to 1 × 105 cells/ml. Either no addition, DMSO, or 250 μM aphidicolin were added to wild type, Jumble, and Couscous. After 24 hr, cells were counted and imaged. (A) Aphidicolin successfully blocked cell division, while the no addition control and DMSO control grew for all conditions. Mean density plotted ±S.D for three technical replicates on the same day as imaged. (B) In wild type cells, no chains were observed in the aphidicolin-treated cells, but they were able to successfully grow chains when cultured either with no addition or only DMSO. For Jumble and Couscous, clumps formed in all conditions. Clumps formed in the presence of aphidicolin appear smaller, perhaps due to the lower cell density of the cultures or the lack of cell division, both of which may contribute to clump size. Scale bar = 20 μm.
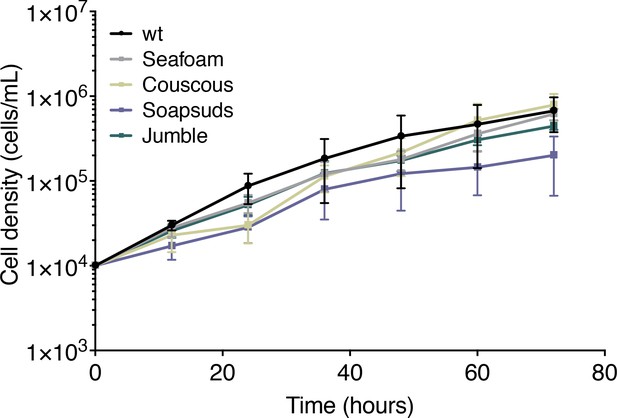
Class C mutant growth curves.
Mutant and wild type cells were plated at a density of 1 × 104 cells/ml and counted every 12 hr to assess growth. Mean density plotted ±SD (n = 2 biological replicates with three technical replicates).
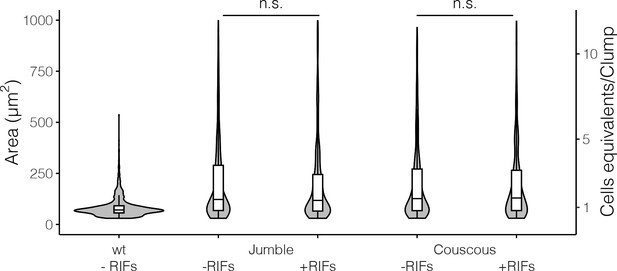
Jumble and Couscous clumps formed in the absence or presence of RIFs are comparable in size.
Jumble and Couscous were cultured for 24 hr, either without RIFs or with RIFs. To perform the clumping assay, cells cultured either with or without RIFs were vortexed and then incubated for 30 min. Wild type cells without RIFs were included as a negative control. Clump sizes were quantified by automated image analysis. Data are presented as violin boxplots, showing the median cell number (horizontal line), interquartile range (white box), and range excluding outliers (vertical line). There were no significant differences in clump size in mutants treated with RIFs or without RIFs (K-S test, n.s. = not significant, p > 0.05).
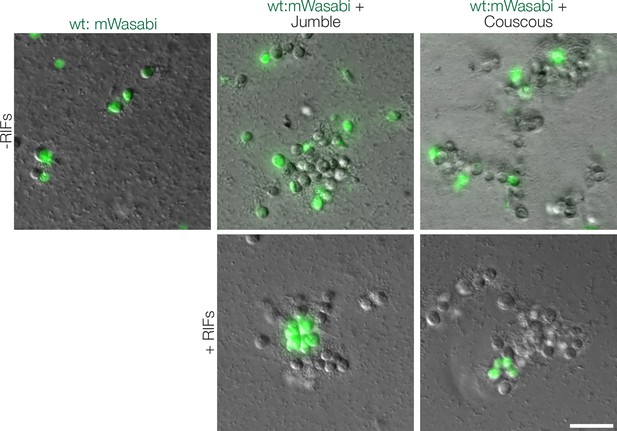
Jumble and Couscous cells adhere to wild type cells.
Fluorescent mWasabi expressing wild type cells uninduced or induced to form rosettes by the addition of RIFs were mixed with either Jumble or Couscous cells and imaged after 30 min. Mutant cells adhered non-specifically to each other and wild type cells. Scale bar = 20 μm.
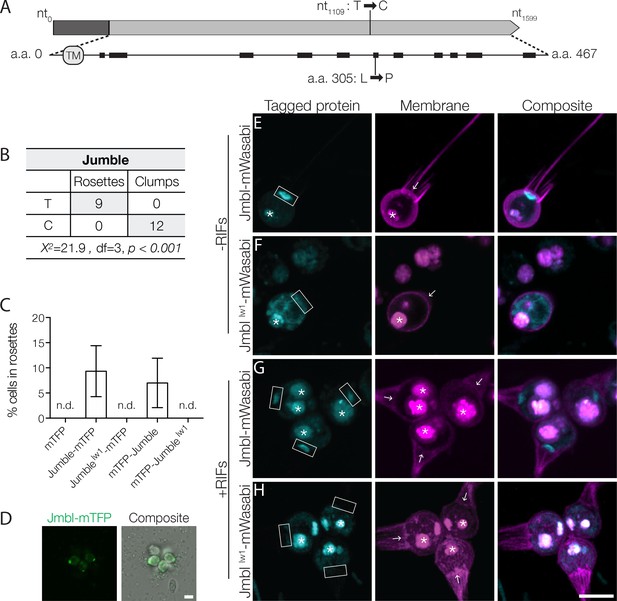
Jumble maps to a predicted glycosyltransferase that localizes to the Golgi apparatus.
(A) Jumble has a predicted transmembrane domain (marked TM) and secondary structure (alpha helices marked by black rectangles). Structural homology algorithms predict that Jumble is structurally related to well-characterized glycosyltransferases (Figure 2—figure supplement 2B). The mutant gene has a T to C mutation at nucleotide 1109 that causes an amino acid substitution of proline to leucine at amino acid position 305. (B) A backcross of a mutant F1 progeny to the Mapping Strain yielded nine rosette-forming F2 isolates with the wild type T allele and twelve clumpy F2 isolates with the jumblelw1 C allele. The inheritance significantly deviated from expected Mendelian inheritance of unlinked traits and confirmed the tight linkage between the jumblelw1 allele and the clumpy, rosetteless phenotype. X2 = Chi squared value, d.f. = degrees of freedom. (C,D) Transgenic expression of jumble-mTFP and mTFP-jumble rescued rosette development in the Jumble mutant, but jumblelw1-mTFP, mTFP-jumblelw1, or mTFP did not. RIFs were added immediately after transfection and 40 μg/ml puromycin was added 24 hr post-transfection to select for transformants. (C) Rosette development was measured as the % of cells in rosettes 72 hr post-transfection and shown as mean ± SD. n.d. = no detected rosettes. (n = 200 cells counted from each of 3 technical replicates; two biological replicates). (D) Rosettes transgenically complemented with jumble-mTFP in the Jumble mutant appeared phenotypically wild type and most cells in rosettes had detectable fluorescent expression at the apical base of the cell. Representative rosette shown. (E–H) To examine localization, Jumble-mWasabi or Jumblelw1-mWasabi (cyan) under the efl promoter were co-expressed with membrane marker-mCherry (magenta) in wild type S. rosetta. Jumble-mWasabi localizes to the apical pole of cells grown (E) without RIFs or (G) with RIFs, consistent with the localization of the Golgi apparatus. When expressed in otherwise wild type cells grown (F) without RIFs or (H) with RIFs, the mutant Jumblelw1-mWasabi incorrectly localizes to the ER and food vacuole. Boxes indicate the inferred location of the Golgi apparatus at the apical pole of the cell. The food vacuole (asterisk) was often visualized due to autofluoresence from ingested bacteria or through accumulation of the fluorescent markers in the food vacuole, perhaps through autophagy. For reference, arrows indicate the base of the flagellum although the flagellum may not be visible in the plane of focus shown. Scale bars = 5 μm.
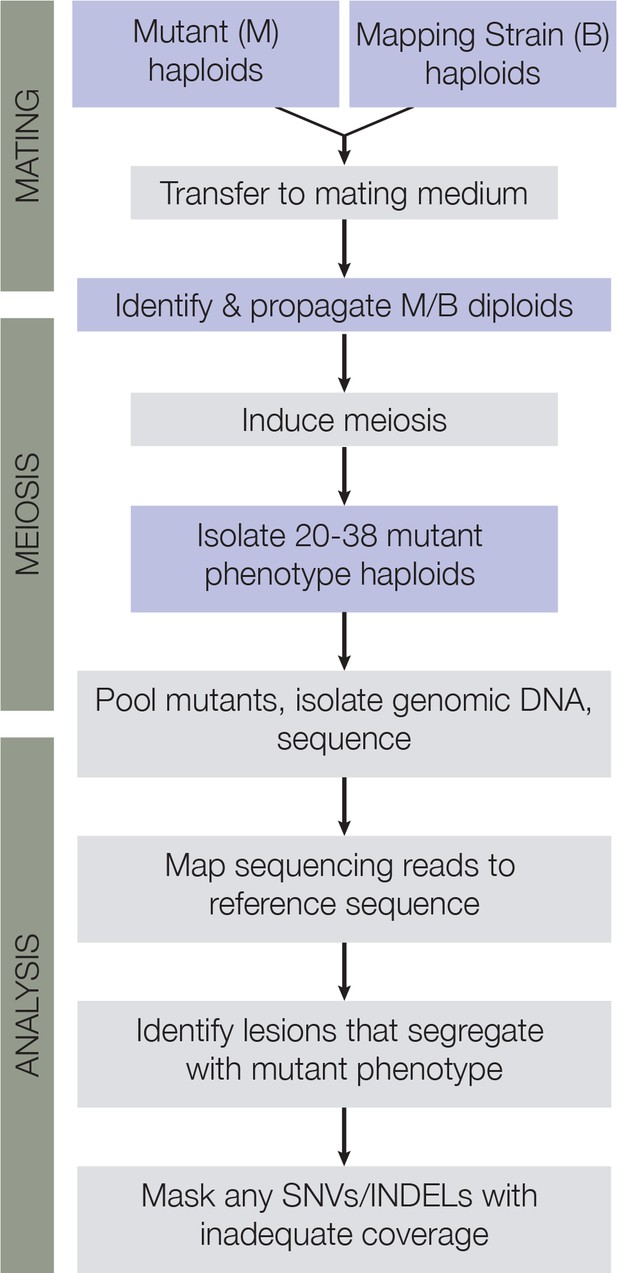
Mapping cross scheme.
Flow chart of the steps used in mapping cross and bulk segregant analysis.
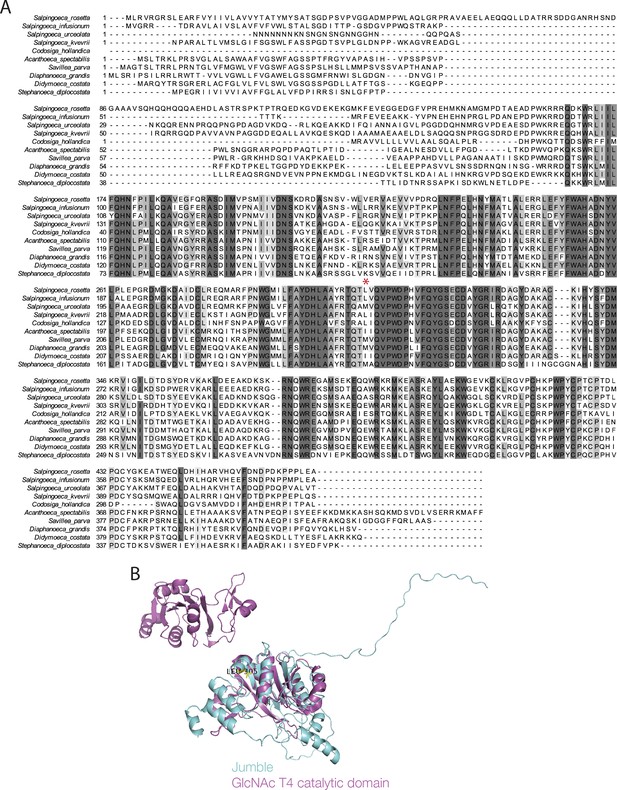
Alignment of Jumble homologs and predicted structure.
(A) S. rosetta Jumble amino acid sequence was aligned to the predicted sequences encoded by homologs from nine other choanoflagellate species, first identified by best reciprocal BLAST using the transcriptomes reported in Richter et al. (2018). Red asterisk indicates the location of the causative mutation in the S. rosetta jumble gene. (B) The structure of Jumble protein predicted by HHphred (teal) was aligned to the catalytic domain of human polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4; purple). The mutated leucine at 305 is found in a predicted alpha helix.

Alignment of Jumble to fungal homologs.
S. rosetta Jumble protein sequence was aligned to predicted/unannotated protein sequences from four fungal species identified by best reciprocal BLAST: Saitoella complicata (NCBI accession XP_019021578.1), Dactylellina haptotyla (NCBI accession EPS43829.1), Naematelia encephala (NCBI accession ORY22834.1), and Tuber magnatum (NCBI accession PWW71609.1).
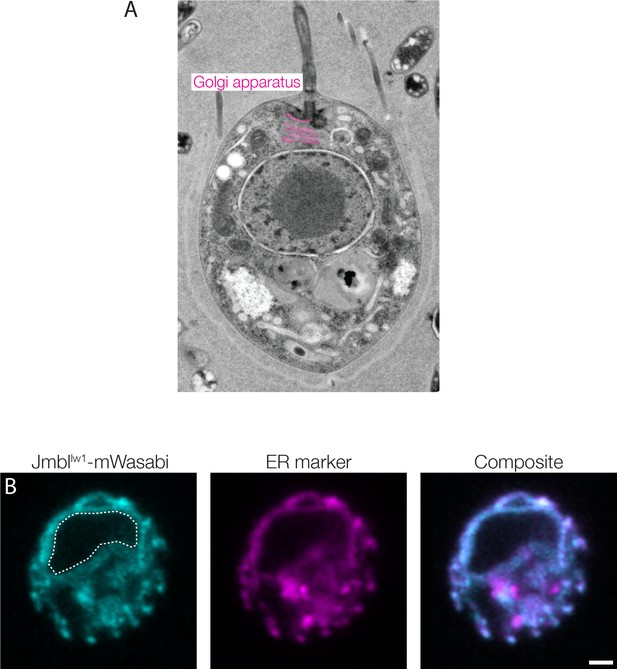
Ultrastructure of S.rosetta and ER co-localization of Jumblelw1.
(A) A transmission electron micrograph shows the ultrastructure of S. rosetta. The Golgi apparatus has been pseudo-colored pink and labelled. Image provided courtesy of Kent McDonald and adapted from Booth et al. (2018). (B) Jumblelw1-mWasabi fusion protein shows partial co-localization with the mCherry-ER marker when expressed in wild type S. rosetta. Dashed line marks the inferred location of the nucleus. Scale bar = 1 μm.
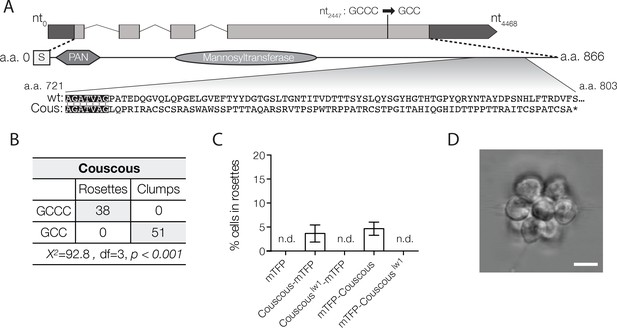
Couscous maps to a predicted mannosyltransferase with a PAN/Apple domain.
(A) Couscous has a predicted signal sequence (S), a PAN/Apple domain (PAN), and a mannosyltransferase domain. The causative lesion is a 1-base pair deletion at nucleotide position 2447 that causes a frameshift at amino acid 728, resulting in 75 amino acids that do not align between the wild type and mutant (Cous) sequences, and an early stop codon (*) at amino acid 803. (B) Independent backcrosses of two individual mutant F1 progeny to the Mapping Strain yielded 38 rosette-forming F2 isolates with the wild type GCCC allele and 51 clumpy F2 isolates with the couscouslw1 GCC allele. The inheritance significantly deviated from expected Mendelian inheritance of unlinked traits and confirmed the tight linkage between the couscouslw1 allele and the clumpy, rosetteless phenotype. X2 = Chi squared value, d.f. = degrees of freedom. (C, D) Rosette formation in Couscous mutant cells can be rescued by transgenic expression of couscous-mTFP or mTFP-couscous, but not couscouslw1-mTFP, mTFP-couscouslw1, or mTFP alone. RIFs were added immediately after transfection and 40 μg/ml puromycin was added 24 hr post-transfection to select for positive transformants. (C) Rosette development (mean ± SD) was measured as the % of cells in rosettes 72 hr after transfection and treatment with RIFs. n.d. = no detected rosettes. (n = 200 cells counted from each of 3 technical replicates; two biological replicates). (D) Rosettes transgenically complemented with couscous-mTFP in the Couscous mutant appeared phenotypically wild type. Representative rosette shown. Scale bar = 5 μm.
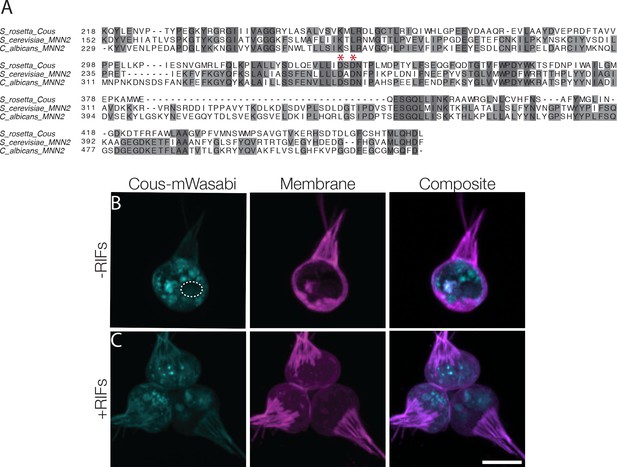
Couscous homology and localization.
(A) The predicted mannosyltransferase domain from S. rosetta was aligned to the alpha-mannosyltransferase domain of MNN2 genes from S. cerevisiae (NCBI accession NP_009571.1) and C. albicans (NCBI accession XP_710276.1). Red asterisks highlight the conserved DXD motif of many glycosyltransferases. (B) The transgenes mCherry-membrane marker and Couscous-mWasabi fusion protein were expressed in wild type S. rosetta. The Couscous fusion localized to puncta distributed throughout the cell (to the exclusion of an unidentified organelle; circle) and faintly at the cell collar. Scale bar = 5 μm.
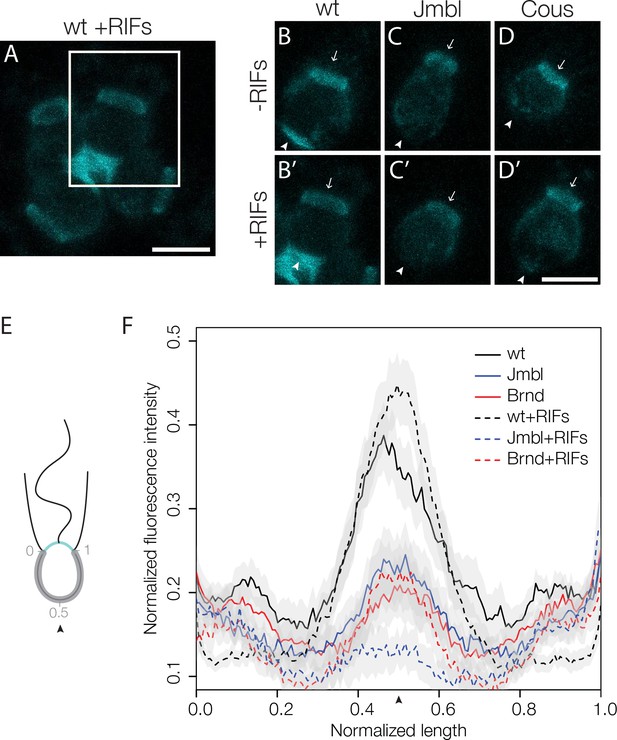
Disruption of basal glycosylation patterns in Jumble and Couscous mutants.
FITC-labelled jacalin binds the apical and basal poles of wild type single cells (B) and becomes enriched in the ECM in the center of rosettes (A, B’ boxed region from A). Although FITC-jacalin staining appeared normal at the apical poles of Jumble (C) and Couscous (D) mutant cells, FITC-jacalin staining at the basal poles of cells was undetectable in cells grown either in the absence (-RIFs; C, D) or presence (+RIFs; C’, D’) of RIFs. Arrows mark the apical pole and arrowheads mark the basal pole. (E) Cartoon depicts how jacalin fluorescence was measured. Starting with micrographs of FITC-jacalin stained cells, a line was drawn tracing from one edge of the collar around the cell body to the other edge of the collar, and the underlying fluorescent signal was normalized for cell size and background intensity. (F) The average normalized fluorescence intensity of jacalin measured in at least 59 cells for each condition was graphed against the normalized length of the cell body (n = 2 biological replicates). Jumble and Couscous -/+RIFs have reduced jacalin binding at the basal pole compared to wild type -/+RIFs. Gray shadows indicate 95% confidence intervals. Scale bars = 5 μm.
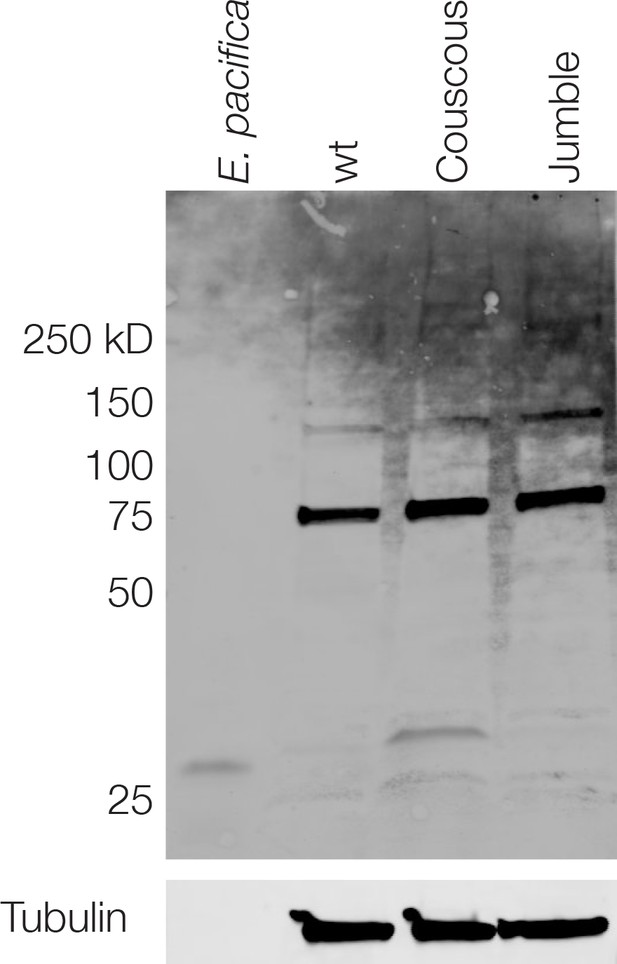
Jacalin Western blot in cell lysates.
Whole cell lysates from E. pacifica (co-cultured prey bacteria), wild type S. rosetta, Couscous, and Jumble were probed with jacalin. No clear differences in banding pattern were observed among the S. rosetta strains, except for a small band ~25 kD in the Couscous lysate that is likely from E. pacifica contamination. Tubulin was probed as a loading control.
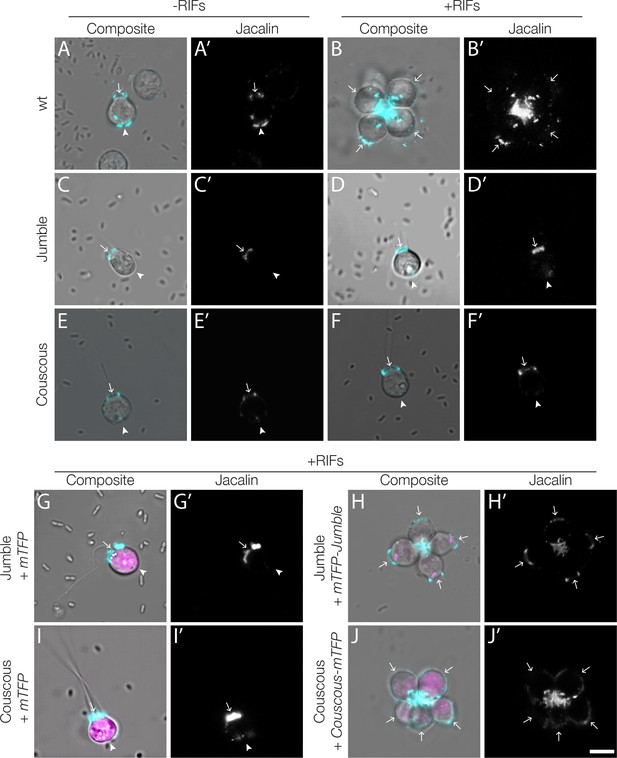
Transgenic rescue restores jacalin staining at the center of complemented rosettes.
(A–F) Biotinylated-jacalin labelled with streptavidin Alexa Fluor 647 has the same localization pattern in the absence (A, C, E) and in the presence (B, D, F) of RIFs as that observed with FITC-labelled jacalin (Figure 4A–D). In wild type cells, jacalin binds the apical and basal poles of single cells (A) and becomes enriched at the center of wild type rosettes (B). In the mutants Jumble (C, D) and Couscous (E, F), jacalin staining was severely reduced at the basal poles both in the absence (C, E) and in the presence (D, F) of RIFs, while the apical pole staining appeared similar to wild type single cells. (G, I) Transfection of Jumble (G) and Couscous (I) with mTFP alone did not restore jacalin localization to the basal pole. Shown here in the presence of RIFs. (H, J) However, Jumble (H) and Couscous (J) complemented with mTFP-jumble or couscous-mTFP, respectively, form rosettes with jacalin localized in the center as observed in wild type rosettes. Arrows mark the apical pole and arrowheads mark the basal pole. Scale bar = 5 μm.
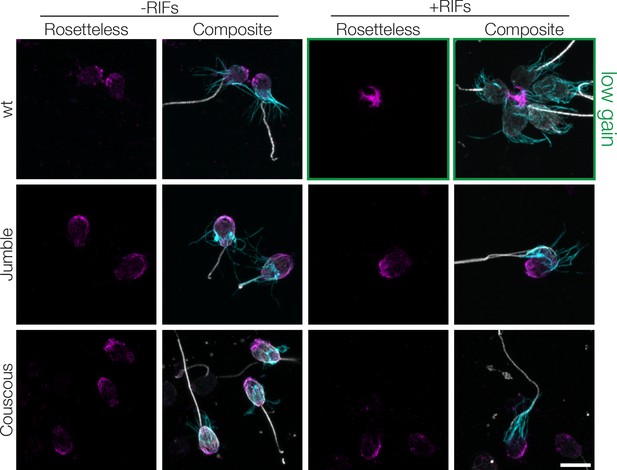
Rosetteless staining in wild type and mutant cells.
Jumble and Couscous cells grown without and with RIFs were stained for Rosetteless (magenta), tubulin (gray), and actin (cyan). In uninduced Jumble and Couscous cells, Rosetteless staining localizes to the basal pole, similar to wild type cells. Following treatment of wild type cells with RIFs, Rosetteless staining becomes highly enriched in the center of rosettes and must be imaged with less gain for clarity (0.3% laser power, gain = 650 indicated by green boxes), while Rosetteless is not enriched or apparently secreted from the basal poles of Jumble or Couscous cells (2% laser power, gain = 800). Scale bar = 20 μm.
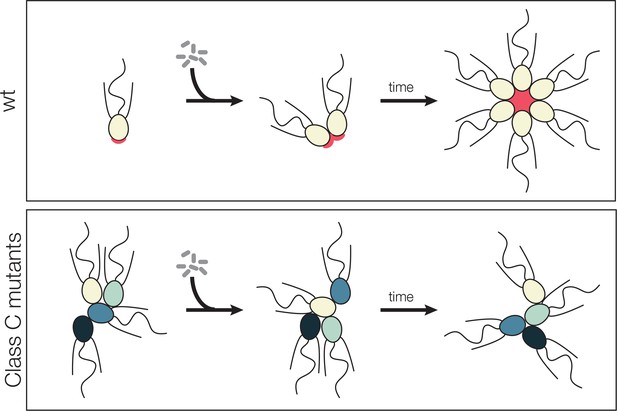
Model for promiscuous clumping in rosette defective Class C mutants.
Wild type S. rosetta has a glycosylated basal patch of ECM (red) as marked by the lectin jacalin that becomes enriched during the course of rosette formation. The Rosetteless protein, required for rosette formation and speculated to play a structural role in holding rosettes together, localizes to the same location on the basal pole of cells and becomes similarly enriched as rosette form. Mutants lack the glycosylated basal patch of jacalin staining. The altered cell surface could lead to clumping, either through mis-regulation of cell adhesion molecules or exposure of a normally masked adhesive cell surface. The same alteration that allows clumping of Class C mutants also prevents rosette development, perhaps by disrupting glycan modification on the Rosetteless protein or one of its interaction partners.
Tables
Phenotypes of wild type and Class C mutants
https://doi.org/10.7554/eLife.41482.007| Strain | % cells in rosettes | Cell interactions | Successful outcross? | |
|---|---|---|---|---|
| wild type | 87.7 | Non-clumping | Yes | |
| Seafoam | 0 | Clumping | No | |
| Soapsuds | 0 | Clumping | No | |
| Couscous | 0 | Clumping | Yes | |
| Jumble | 0 | Clumping | Yes | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | |
|---|---|---|---|---|---|
| Gene (Salpingoeca rosetta) | jumble | NA | GenBank accession EGD72416/NCBI accession XM_004998928; | ||
| Gene (S. rosetta) | couscous | NA | GenBank accession EGD77026/NCBI accession XM_004990809 | ||
| Strain, strain background (S. rosetta) | wt | PMID: 24139741 | ATCC PRA-390; accession number SRX365844 | ||
| Strain, strain background (S. rosetta) | Mapping Strain | PMID: 24139741 | accession numberSRX365839 | ||
| Strain, strain background (S. rosetta) | Jumble | PMID: 25299189 | accession number SRR7866767 | ||
| Strain, strain background (S. rosetta) | Couscous | PMID: 25299189 | accession number SRR7866768 | Previously named Branched | |
| Strain, strain background (S. rosetta) | Seafoam | PMID: 25299189 | accession number SRR8263910 | ||
| Strain, strain background (S. rosetta) | Soapsuds | PMID: 25299189 | accession number SRR8263909 | ||
| Strain, strain background (Algoriphagus macihipongenesis) | Algoriphagus macihipongenesis | PMID: 22368173 | ATCC BAA-2233 | ||
| Strain, strain background (Echinicola pacifica) | Echinicola pacifica | PMID: 16627637 | DSM 19836 | ||
| Strain, strain background (Vibrio fishceri) | Vibrio fishceri ES114 | PMID: 15703294 | ATCC 700601 | ||
| Antibody | anti-Rosetteless | PMID: 25299189 | (1:400) | ||
| Recombinant DNA reagent | mCherry plasma membrane marker | PMID: 30281390 | RRID:Addgene_109094; Addgene ID NK624 | ||
| Recombinant DNA reagent | mCherry ER marker | PMID: 30281390 | RRID:Addgene_109096; Addgene ID NK644 | ||
| Recombinant DNA reagent | pEFl5'-Actin3'::jumble-mWasabi | this paper | Addgene ID NK690 | pUC19 backbone with 5’ S. rosetta elongation factor L (efl) promoter, jumble, mWasabi, and 3’ UTR from actin; assembled by Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::jumblelw1-mWasabi | this paper | Addgene ID NK691 | pUC19 backbone with 5’S. rosetta elongation factor L (efl) promoter, jumblelw1, mWasabi, and 3’ UTR from actin; assembled by Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::couscous-mWasabi | this paper | Addgene ID NK692 | pUC19 backbone with 5’ S. rosetta elongation factor L (efl) promoter, couscous, mWasabi, and 3’ UTR from actin; assembled by Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-mTFP | this paper | Addgene ID NK676 | pUC19 backbone with 5’ S. rosetta elongation factor L (efl) promoter, S. rosetta codon optimized puromycin resistance gene (pac), mTFP, and 3’ UTR from actin; assembled by Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-jumble-mTFP | this paper | Addgene ID NK694 | Parent vector: pEFl5'- Actin3'::pac-P2A-mTFP; jumble inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-jumblelw1-mTFP | this paper | Addgene ID NK695 | Parent vector: pEFl5'- Actin3'::pac-P2A-mTFP; jumblelw1 inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-mTFP-jumble | this paper | Addgene ID NK696 | Parent vector: pEFl5'- Actin3'::pac-P2A-mTFP; jumble inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-mTFP-jumblelw1 | this paper | Addgene ID NK697 | Parent vector: pEFl5'- Actin3'::pac-P2A-mTFP; jumblelw1 inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-couscous-mTFP, | this paper | Addgene ID NK698 | Parent vector: pEFl5'- Actin3'::pac-P2A-mTFP; couscous inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-couscouslw1-mTFP | this paper | Addgene ID NK699 | Parent vector: pEFl5'- Actin3'::pac-P2A-mTFP; couscouslw1 inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-mTFP-couscous | this paper | Addgene ID NK700 | Parent vector: pEFl5'-Actin3':: pac-P2A-mTFP; couscous inserted using Gibson assembly | |
| Recombinant DNA reagent | pEFl5'-Actin3'::pac-P2A-mTFP-couscouslw1 | this paper | Addgene ID NK701 | Parent vector: pEFl5'-Actin3':: pac-P2A-mTFP; couscouslw1 inserted using Gibson assembly | |
| Other | FITC-labelled jacalin | Vector Labs | RRID:AB_2336460; Vector Labs: Cat. No.FLK-4100 | (1:400) | |
| Other | biotinylated jacalin | Vector Labs | RRID:AB_2336541; Vector Labs: Cat. No. B-1155 | ||
| Other | Streptavidin Alexa Fluor 647 conjugate | Thermo Fisher Scientific | Thermo Fisher Scientific: Cat. No. 32357 | ||
Additional files
-
Supplementary file 1
Supplementary tables.
(1) Table S1. Phenotypic classes of mutants isolated in this study and in the Levin et al. (2014) screen. (2) Table S2. Segregating variants in Rosetteless mapping cross. (3) Table S3. Segregating variants in Jumble mapping cross. (4) Table S4. Segregating variants in Couscous mapping cross. (5) Table S5. Fluorescent lectins tested.
- https://doi.org/10.7554/eLife.41482.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41482.021





