Molecular basis for activation of lecithin:cholesterol acyltransferase by a compound that increases HDL cholesterol
Figures
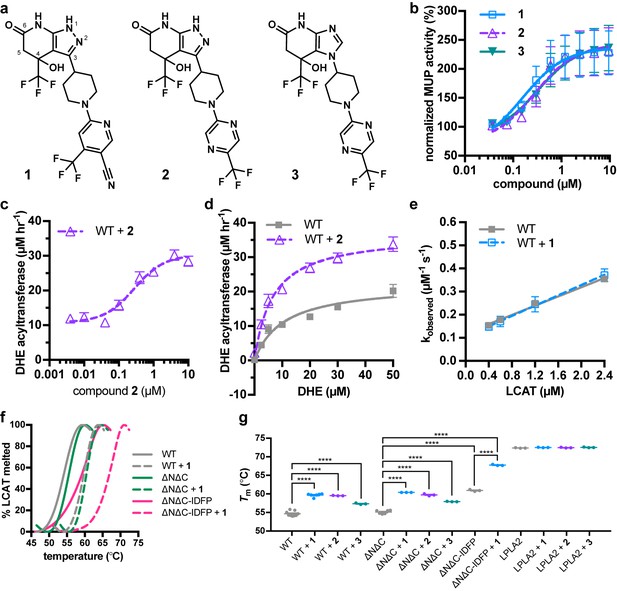
Piperidinylpyrazolopyridine and related activators stimulate and stabilize LCAT.
(a) Structure of compounds 1 (patent example 95 (Kobayashi et al., 2015a)), 2 (patent example 46 (Kobayashi et al., 2015a)), and 3 (patent example 3 (Onoda et al., 2015)). (b) All three activators stimulate LCAT in a micelle-based MUP assay. Data shown are mean ± s.e.m. from three independent experiments, and data were normalized to basal LCAT activity. (c) Titration of compound 2 (used in this particular assay due to its lower background fluorescence) in the DHE acyltransferase assay. Data shown are mean ± s.e.m. from three independent experiments performed in triplicate. (d) The addition of 5 μM compound 2 stimulates LCAT acyltransferase activity. Data shown are mean ± s.e.m. from three independent experiments performed in triplicate. (e) The addition of 10 μM compound 1 does not affect LCAT binding to HDL as measured with BLI. Plot used to determine kon, koff, and hence Kd. Data are mean ± s.e.m. of three independent experiments. (f) Representative DSF data highlighting the additive increase in Tm induced by combination of 1 and IDFP. Data are normalized from 0% to 100% using the lowest and highest values, respectively. (g) Compounds 1, 2, and 3 stabilize WT, ∆N∆C, and ∆N∆C-IDFP LCAT, but not LPLA2. DSF data are mean ± s.e.m. of at least three independent experiments performed in duplicate. ****p<0.0001 by one-way analysis of variance followed by Tukey’s multiple comparisons post-test. Each protein without ligand was compared to same variant with ligand, and non-significant pairs are not shown. WT compared to ∆N∆C was not significant.
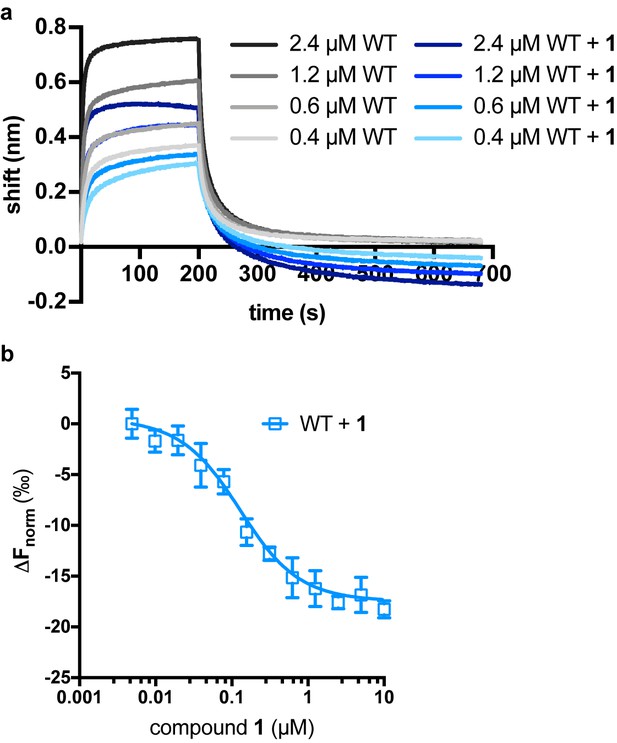
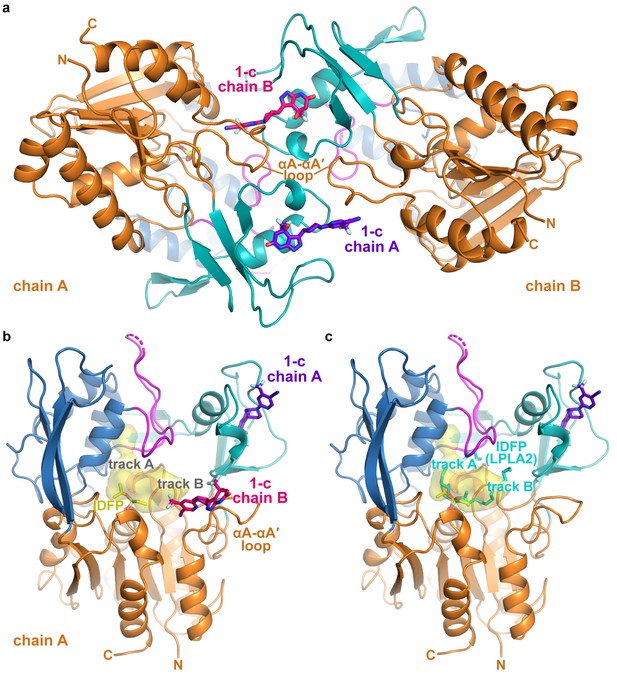
Effects of LCAT binding to compound 1.
(a) LCAT analyzed with ApoA-I HDLs at different concentrations in order to determine Kd. Raw data with WT LCAT is shown in black to gray, whereas WT in complex with compound 1 is shown in navy to light blue. (b) Microscale thermophoresis (MST) data for compound 1 binding to LCAT with a Kd value of 100 ± 14 nM.
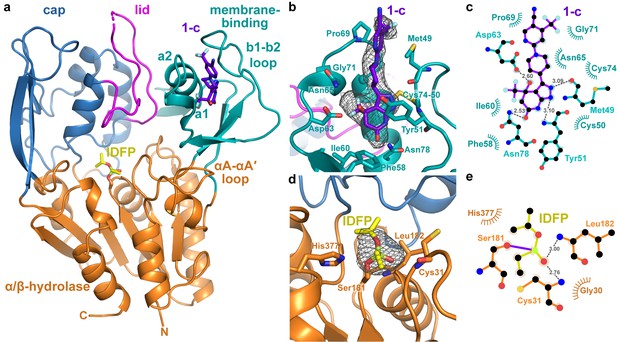
Structure of the ΔNΔC-IDFP·1 complex.
(a) 3.1 Å X-ray crystal structure highlighting the three domains of LCAT and the binding sites for compound 1-c (purple) and IDFP (yellow), shown as stick models. The hydrolase domain is shown in orange, cap domain in blue, lid in magenta, and membrane-binding domain (MBD) in teal. (b) Closeup of 1-c bound to the MBD, with |Fo|-|Fc| omit map density contoured at 3 σ in gray mesh. (c) LigPlot (Laskowski and Swindells, 2011) of 1-c bound to LCAT showing interactions between protein and ligand. Hydrogen bonds are indicated by gray dashed lines with distances in Å. (d) IDFP attached to catalytic Ser181, with |Fo|-|Fc| omit map density contoured at 3 σ in gray mesh. (e) LigPlot of IDFP bound covalently to LCAT at Ser181. The covalent point of attachment is indicated by a purple bond. Protein carbons are colored according to their respective domains or ligands (panel a), whereas nitrogens are blue, oxygens red, sulfurs yellow, and phosphate lime green.
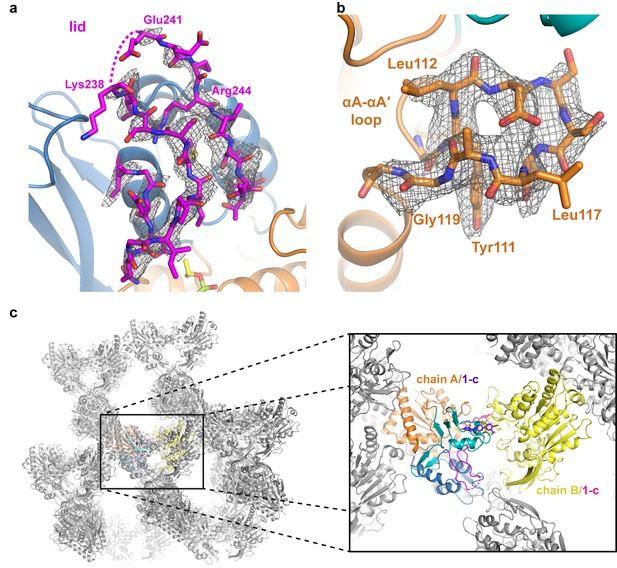
Electron density maps and crystal packing of the ΔNΔC-IDFP·1 complex.
(a–b) |Fo|-|Fc| omit map density contoured at 2.5 σ in gray mesh revealing (a) the lid and (b) the αA-αA′ loop of chain A. In (a) the dashed line indicates disordered residues. (c) Crystal packing observed for the ΔNΔC-IDFP·1 complex with symmetry mates in grey that are within 50 Å of the structure. On right, the zoomed in slice highlights interactions with the asymmetric unit.
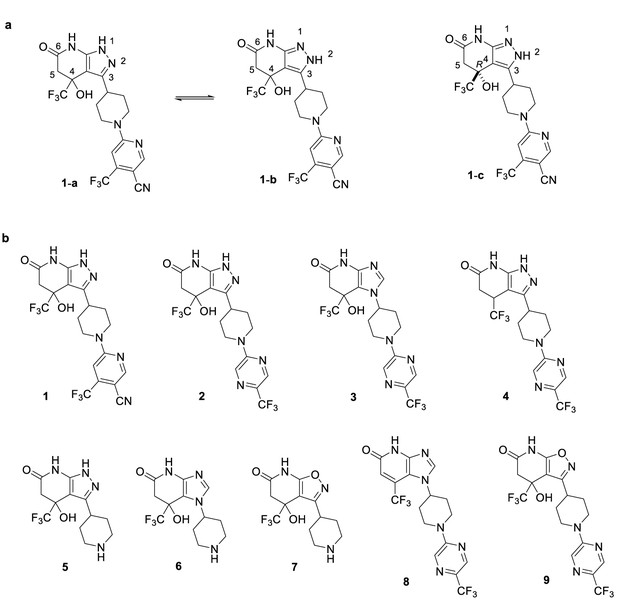
Compound structures and numbers.
(a) Compounds 1-a and 1-b are tautomers of 1, where 1-b is the dominant isoform in the co-crystal structure. Compound 1 has a stereocenter at the C4 position and was synthesized as a racemic mixture, however the binding site is only compatible with the R enantiomer, 1-c. (b) Structure of compounds 1 (patent example 95 (Kobayashi et al., 2015a)), 2 (patent example 46 (Kobayashi et al., 2015a)), and 3 (patent example 3 (Onoda et al., 2015)) are the key compounds synthesized and examined in this study. Compound 4 (patent example 10 (Kobayashi et al., 2015b)) is referenced in the text. Compounds 5–7 are the head groups of related compounds that were synthesized as intermediates, and 8 and 9 were synthesized as part of a structure-activity analysis.
Asymmetric unit of the ΔNΔC-IDFP·1 crystals and interactions of compound 1-c.
(a) Compound 1-c helps to bridge the two LCAT chains to form a pseudo 2-fold interface. (b) LCAT from chain A is shown with compound 1-c from both chain A (bound to membrane binding domain, purple) and chain B (docking to track B in the active site, pink). (c) For comparison, IDFP from the LPLA2-IDFP structure (PDB entry 4X91 chain A (Glukhova et al., 2015)) is shown with cyan carbons after alignment of LPLA2 and LCAT. IDFP adopts two different orientations in 4X91, thus revealing two potential tracks for acyl chains. The structure shown is of ΔNΔC-IDFP·1 with bound IDFP (yellow) and 1-c (purple). The LCAT substrate binding surface is highlighted in yellow. The interior surface showing track A was created using HOLLOW (Ho and Gruswitz, 2008) and rendered in PyMOL. The surface as shown only extends partially into track B because it is designed to show interior surfaces, and track B is more exposed to solvent.
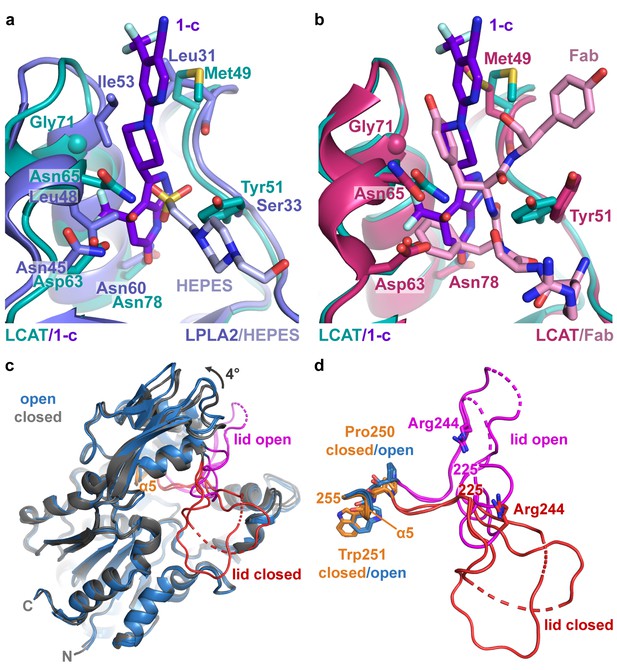
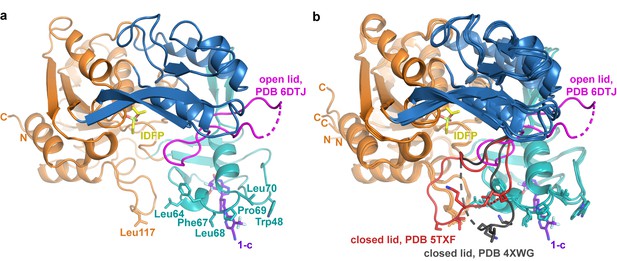
Comparison of LCAT and LPLA2 structures.
(a) ΔNΔC-IDFP·1 structure aligned with LPLA2 (blue, PDB entry 4X90) bound to HEPES (light blue). Residues that are not conserved within the binding pocket are labeled and shown as stick models. (b) Structure of ΔNΔC-IDFP·1 aligned to that of 27C3–LCAT–Fab1 (dark pink, PDB entry 5BV7 with Fab1 shown in pink), highlighting residues that adjust conformation to accommodate the different ligands. (c) Four LCAT crystal structures aligned to show differences between the open and closed states. Closed (presumably inactive) structures are shown in gray (PDB entries 4XWG and 5TXF) with orange hinge and red lid. Open structures (structure reported here and 27C3–LCAT–Fab1) are shown in blue with magenta lid. Dashed lines indicate disordered residues. (d) Close up of structures from (c) only depicting the lid and hinge region. Hinge residues Pro250 and Trp251 and lid residue Arg244 are shown as stick models.
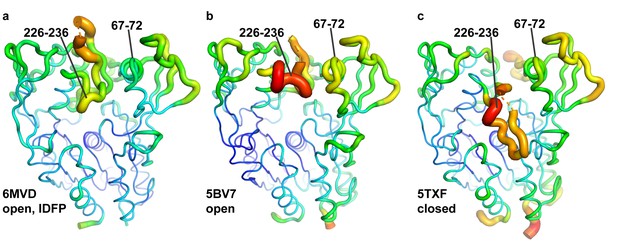
The ΔNΔC-IDFP·1 structure has lower temperature factors in the membrane binding domain and lid.
(a) ΔNΔC-IDFP·1 open structure, (b) 27C3–LCAT–Fab1 open structure (Gunawardane et al., 2016), and (c) LCAT-closed structure (Manthei et al., 2017) are shown using B-factor putty representation in PyMol. The blue to green coloring and small tube width indicates lower B-factors whereas the yellow to red coloring and wide tube indicates higher B-factors. Residues 67–72 and 226–236 have lower B-factors in the ΔNΔC-IDFP·1 structure ()and also had lower hydrogen-deuterium exchange (HDX) in the presence of IDFP (Manthei et al., 2017).
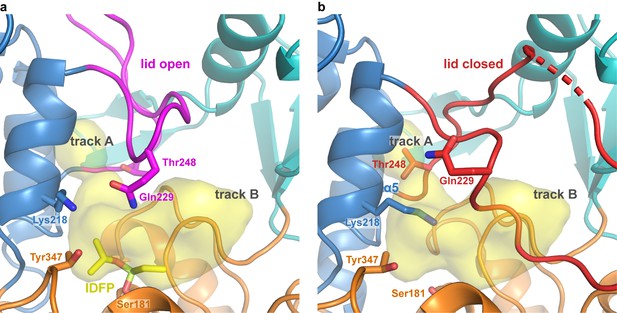
Hinge and lid movement modulate lipid binding tracks.
(a) When the lid is open, both tracks are available for acyl chain binding, and residues Lys218, Gln229, and Tyr347 are in position to coordinate phosphate in the lipid head group. The structure shown is of ΔNΔC-IDFP·1 with bound IDFP (yellow). The LCAT substrate binding surface is highlighted in yellow. The interior surface showing track A was created using HOLLOW (Ho and Gruswitz, 2008) and rendered in PyMOL. The surface as shown only extends partially into track B because it is designed to show interior surfaces, and track B is more exposed to solvent. (b) When the lid is closed, track A becomes blocked by the hinge and lid movement, and changes in the position of the α5 helix causes Thr248 to block the back of track A. Lys218 also occludes part of the binding site and portions of the lid pack in track B. The structure shown is LCAT-closed (PDB code 5TXF (Manthei et al., 2017)).
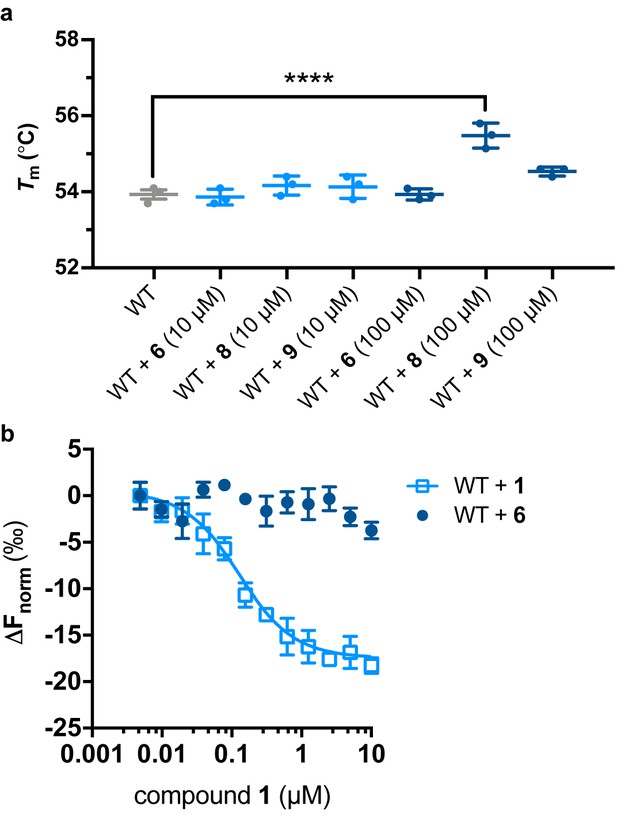
Structure-activity relationships.
(a) Compounds 6, 8, and 9 do not stabilize WT LCAT at 10 μM, but 8 does at 100 μM. Differential scanning fluorescent (DSF) data are mean ± s.e.m. of at least three independent experiments performed in duplicate. ****p<0.0001 by one-way analysis of variance followed by Tukey’s multiple comparisons post-test. WT LCAT without ligand was compared to WT with compound and non-significant comparisons are not shown. (b) MST data showing that compound 6 does not bind to WT LCAT, whereas compound 1 does.
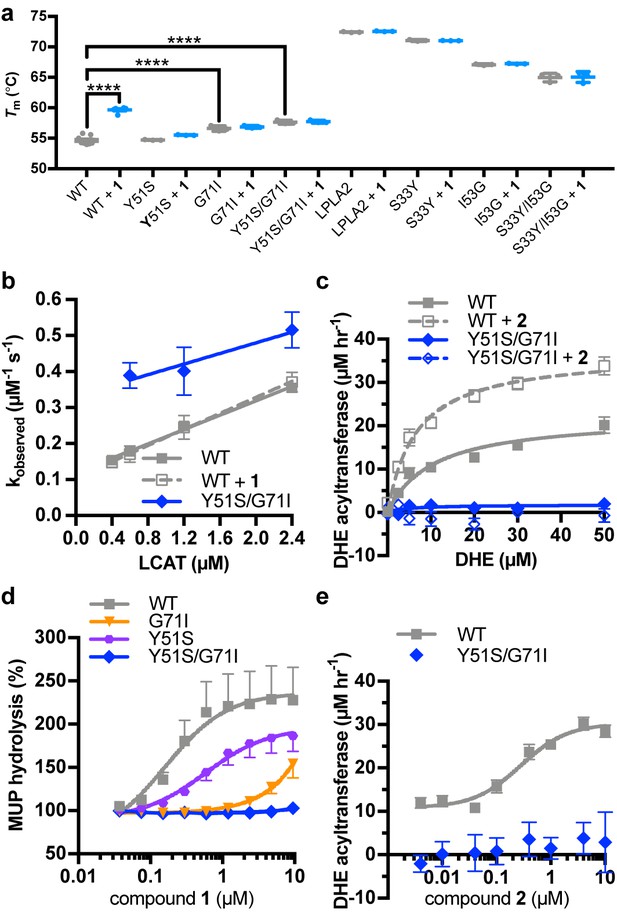
Characterization of activator binding site mutants.
(a) Perturbation of the activator binding site leads to loss of responsiveness to compound 1, although the G71I and Y51S/G71I variants are themselves stabilized compared to WT LCAT. LPLA2 variants, however, do not bind to 1, and chimeric swaps are destabilized. Data are mean ± s.e.m. of at least three independent experiments performed in duplicate. ****p<0.0001 by one-way analysis of variance followed by Tukey’s multiple comparisons post-test. Each protein without ligand was compared to that same variant with compound 1, and WT LCAT was compared to each LCAT variant. Non-significant comparisons are not shown. (b) Plot used to determine kon, koff, and hence Kd from BLI data for LCAT binding to HDL. Data are mean ± s.e.m. of three independent experiments. (c) DHE acyltransferase assay with peptide HDLs comparing the absence (solid lines) and presence (dashed lines) of 5 μM compound 2, which was used in this assay instead of 1 due to its lower background fluorescence. Data are mean ± s.e.m. of three independent experiments performed in triplicate. (d) Titration of compound 1 in the MUP hydrolysis assay. Data were normalized to basal activity of 100% for each variant to give percent activation. Data are mean ± s.e.m. of three independent experiments. (e) Titration of compound 2 in the DHE acyltransferase assay. Data are mean ± s.e.m. of three independent experiments performed in triplicate.
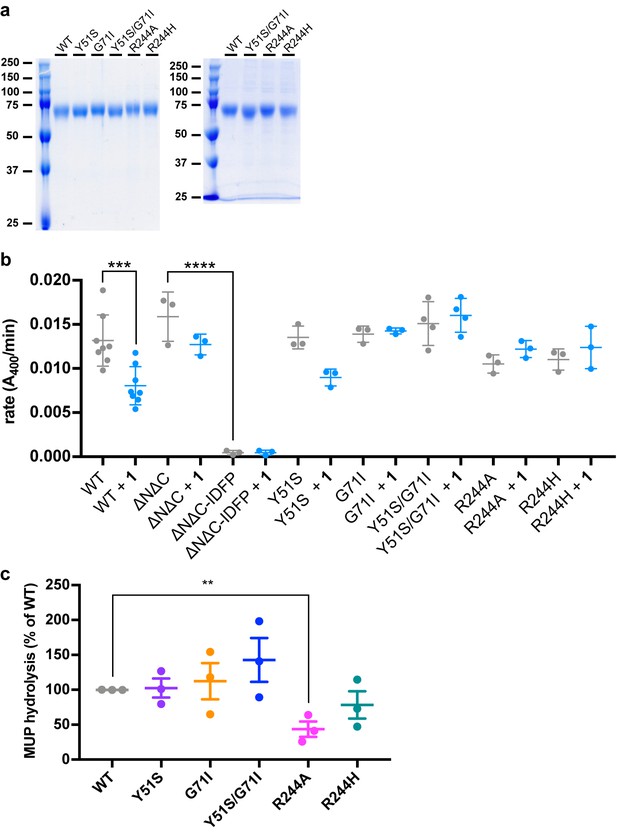
Biochemical characterization of LCAT variants.
(a) SDS-PAGE gels of purified LCAT variants. Left panel shows representative variants used in pNPB, MUP, and DSF experiments that were polished via Superdex 75. Right panel shows representative variants used for DHE acyltransferase and BLI assays. Approximately 1.5 µg of each purified LCAT variant was loaded in each lane. (b) Rates of pNPB hydrolysis for LCAT variants. Data are mean ± s.e.m. of at least three independent experiments. ***p=0.0004, ****p<0.0001 by one-way analysis of variance followed by Tukey’s multiple comparisons post-test. Each protein without ligand was compared to that same variant with ligand, and WT LCAT was compared to each LCAT variant. Non-significant comparisons are not shown. (c) Comparison of basal MUP hydrolysis. Data are mean ± s.e.m. of three independent experiments performed with ≥27 repeats. **p=0.0070 by a two-tailed unpaired t test. WT was compared to each LCAT variant and non-significant comparisons are not shown.
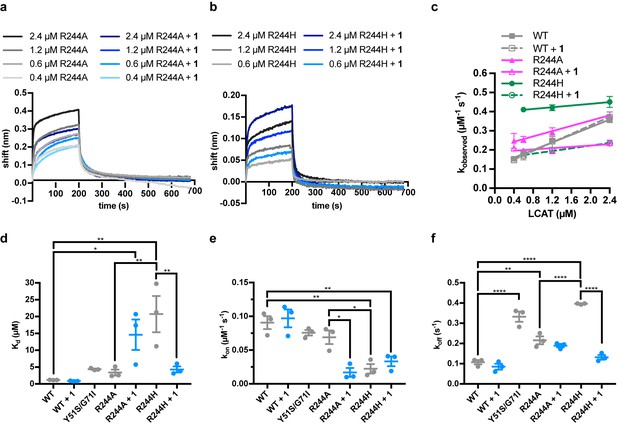
Representative BLI data for LCAT-Arg244 variants.
(a) LCAT analyzed with ApoA-I HDLs at different concentrations in order to determine Kd. R244A is in black to gray, whereas R244A with compound 1 is in navy to light blue. (b) Same as in (a) but with R244H. (c) Plot used to determine kon (slope of fit line), koff (y-intercept), and hence Kd for LCAT binding to HDL. Data are mean ± s.e.m. of three independent experiments. (d) – (f) Individual Kd (d), koff (e), and kon (f) values were calculated for each experiment. Data are mean ± s.e.m. of three independent experiments. * 0.01 <P < 0.05, ** 0.001 <P < 0.01, *** 0.0001 < P < 0.001, ****p<0.0001 by one-way analysis of variance followed by Tukey’s multiple comparisons post-test. Each protein without ligand was compared to that same variant with 1, and WT LCAT was compared to each dataset. R244A was also compared to R244H. Non-significant comparisons are not shown.
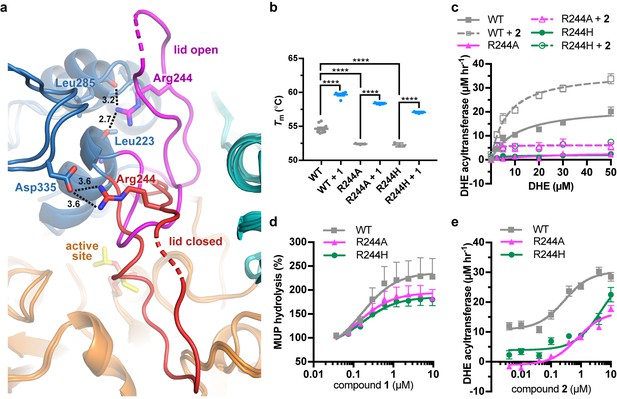
LCAT-Arg244 variants can be partially rescued by LCAT activators.
(a) LCAT-Arg244 acts as part of a molecular switch that interacts with the backbone carbonyls of Leu223 and Leu285 in activated structures of LCAT (magenta lid). In an inactive structure (red lid, PDB entry 5TXF), Arg244 instead interacts with the side chain of Asp335. Hydrogen bonds are indicated by black dashed lines with distances in Å. (b) The Arg244 variants have lower Tm values relative to WT, yet compound 1 can stabilize each to the same extent. Data are mean ± s.e.m. of at least three independent experiments performed in duplicate. ****p<0.0001 by one-way analysis of variance followed by Tukey’s multiple comparisons post-test. (d) DHE acyltransferase assay with peptide-based HDLs comparing the absence (solid lines) and presence (dashed lines) of 5 μM 2. Data are mean ± s.e.m. of three independent experiments performed in triplicate. (c) Titration of 1 in the MUP esterase assay. Data were normalized to basal activity of 100% for each variant to give percent activation. Data are mean ± s.e.m. of three independent experiments. (d) Titration of compound 2 in the DHE acyltransferase assay. Data are mean ± s.e.m. of three independent experiments performed in triplicate.
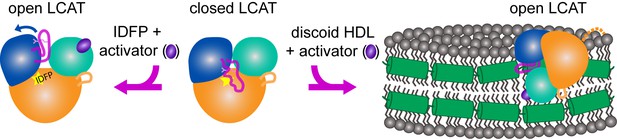
Model of LCAT activation.
We previously described the closed lid structure of LCAT (middle) wherein the lid (magenta coil) would shield the hydrophobic active site of LCAT in solution. In the crystal structure described herein, LCAT adopts an open conformation (left), which is stabilized by IDFP bound in the active site (yellow star) and the small molecule activator (purple ellipse). We hypothesize that LCAT binds to discoid HDL with a similar open lid conformation and that the activator facilitates lipid transport into the active site. Thus the hydrophobic interface is shown binding to the hydrophobic acyl chains on the side of discoidal HDL, and the expected ApoA-I (green helices) interaction is shown as a contact with the lid, providing a structural explanation for ApoA-I activation of LCAT (Cooke et al., 2018). LCAT is depicted with the α/β-hydrolase domain in orange, the cap domain in blue, and the membrane binding domain in teal. The R244 side chain in the lid is shown as pink sticks. The dashed orange line in the HDL complex depicts the disordered N-terminus of LCAT which is also critical for HDL binding.
The activator molecule contributes to a hydrophobic surface.
(a) ΔNΔC-IDFP·1 oriented so that hydrophobic residues from the membrane binding domain and αA-αA′ loop are all in a plane along bottom of the panel. The pyrazine ring of 1-c (purple sticks) also contributes to this plane. The active site lid is shown in magenta. (b) When the lid closes, this interface is partially blocked. The lid from PDB entry 5TXF is shown in red (Manthei et al., 2017), and that of PDB entry 4XWG is gray (Piper et al., 2015).
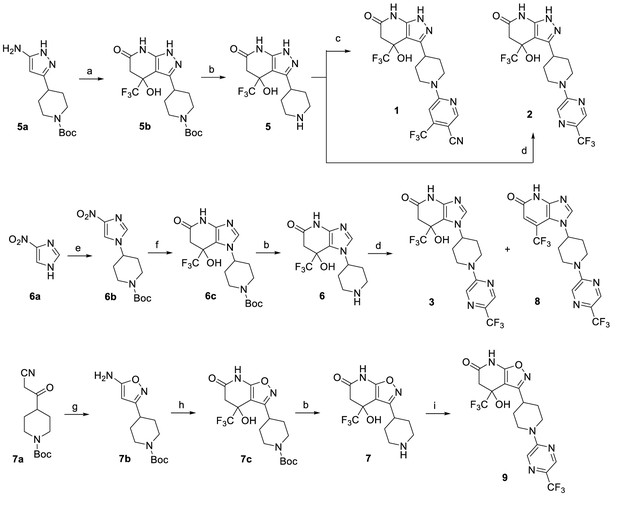
Synthesis of piperidinylpyrazolopyridine and related compounds.
Reagents and conditions: (a) ethyl 4,4,4-trifluoro-3-oxobutanoate, AcOH, 60°C, 3 hr, 57%. (b) HCl (4 M in 1,4-dioxane), 1,4-dioxane, 0°C to RT, 2 hr, 5 (96%), 6 (96%), 7 (83%). (c) 6-chloro-4-(trifluoromethyl)nicotinonitrile, Et3N, EtOH, RT, 1 hr, 13%. (d) 2-chloro-5-(trifluoromethyl)pyrazine, (i-Pr)2NEt, DMSO, RT, 3 hr, 2 (76%), 3 (76%), 8 (~7%). (e) tert-butyl 4-((methylsulfonyl)oxy)piperidine-1-carboxylate, K2CO3, DMF, 110°C, overnight, 38%. (f) H2 balloon, cat. Pd/C, EtOH, RT, 2.5 hr; then ethyl 4,4,4-trifluoro-3-oxobutanoate, EtOH/AcOH (~1:2), 65–70°C, 2.5 hr, 86%. (g) hydroxylamine HCl salt, Et3N, CH2Cl2, sealed, 55°C, overnight, 88%. (h) ethyl 4,4,4-trifluoro-3-oxobutanoate, EtOH/AcOH (~1:2), 70°C, 6 hr, 64%. (i) 2-chloro-5-(trifluoromethyl)pyrazine, Et3N, DMF, RT, 3 hr, 47%.
Videos
Transition between closed and open conformations of LCAT.
The video highlights the opening of the lid and corresponding cap domain movements that occur upon LCAT activation. Arg244 and the residues it interacts with in each conformation, as well as the active site location Ser181 are shown as sticks. Chimera (Pettersen et al., 2004) was used to morph from the closed structure (PDB entry 5TXF (Manthei et al., 2017)) to the activator structure. The video was rendered using PyMOL.
Movement corresponding to the hinge region.
The same morph as depicted in Video 1, but zoomed in on the lid and hinge region. The closed (presumably inactive) structure (PDB entry 5TXF (Manthei et al., 2017)) is shown with orange hinge and red lid. The ∆N∆C-IDFP·1 structure is shown in blue with magenta lid, which is retained during the morph. Dashed lines indicate disordered residues. Hinge residues Pro250 and Trp251 are shown as stick models, as well as the side chain of Arg244 in the lid region. The position of the Cα atom of Gly230 is indicated with a sphere.
Tables
EC50 values of LCAT variants in esterase and acyltransferase assays.
https://doi.org/10.7554/eLife.41604.005| MUP assay EC50 (μM) | DHE assay EC50 (μM) | |||||
|---|---|---|---|---|---|---|
| Variant\Compound | 1 | 2 | 3 | 2 | ||
| WT | 0.16 ± 0.01 | 0.28 ± 0.04 | 0.32 ± 0.05 | 0.28 ± 0.09 | ||
| Y51S | 0.59 ± 0.03 | 0.74 ± 0.2 | 1.6 ± 0.4 | ND | ||
| G71I | >5 | >5 | >5 | ND | ||
| Y51S/G71I | no effect | no effect | no effect | no effect | ||
| R244A | 0.13 ± 0.02 | 0.27 ± 0.04 | 0.40 ± 0.03 | 0.76 ± 0.2 | ||
| R244H | 0.16 ± 0.03 | 0.32 ± 0.03 | 0.47 ± 0.05 | 4.6 ± 2 | ||
-
ND = not determined. In the MUP esterase assay, compound was titrated from 0.04 to 9.5 μM, and reactions were performed in triplicate. In the DHE acyltransferase assay, compound 2 was titrated from 0.004 to 10 μM and reactions were performed three times in triplicate. Values reported are mean ± s.e.m.
Fold activation for LCAT variants in the MUP esterase assay.
https://doi.org/10.7554/eLife.41604.006| Fold activation | |||||||
|---|---|---|---|---|---|---|---|
| Variant\Compound | 1 | 2 | 3 | 6 | 8 | 9 | |
| WT | 2.3 ± 0.4 | 2.3 ± 0.4 | 2.4 ± 0.4 | no effect | 3.7 ± 0.9 | 1.6 ± 0.2 | |
| Y51S | 1.9 ± 0.2 | 1.8 ± 0.1 | 1.9 ± 0.2 | no effect | 2.8 ± 0.6 | 1.1 ± 0.07 | |
| G71I | 1.5 ± 0.4 | 1.7 ± 0.2 | 1.5 ± 0.2 | no effect | 1.2 ± 0.1 | 0.96 ± 0.01 | |
| Y51S/G71I | 1.3 ± 0.3 | 0.99 ± 0.03 | 1.1 ± 0.06 | no effect | 1.1 ± 0.07 | 0.97 ± 0.003 | |
| R244A | 1.7 ± 0.4 | 1.9 ± 0.2 | 1.9 ± 0.2 | no effect | 3.2 ± 0.8 | 1.2 ± 0.1 | |
| R244H | 1.6 ± 0.3 | 1.8 ± 0.1 | 1.8 ± 0.1 | no effect | 2.8 ± 0.6 | 1.3 ± 0.06 | |
-
Compound was titrated from 0.04 to 9.5 μM, and reactions were performed in triplicate with values reported as mean ± s.e.m.
Effect of LCAT mutations and compound 1 on HDL binding.
https://doi.org/10.7554/eLife.41604.007| Variant | kon (s−1 μM−1) | koff (s−1) | Kd (μM) | ||||
|---|---|---|---|---|---|---|---|
| WT | 0.10 ± 0.006 | 0.12 ± 0.008 | 1.2 | ||||
| WT + 1 | 0.11 ± 0.003 | 0.11 ± 0.004 | 1.0 | ||||
| Y51S/G71I | 0.074 ± 0.02 | 0.33 ± 0.03 | 4.5 | ||||
| R244A | 0.069 ± 0.003 | 0.22 ± 0.005 | 3.2 | ||||
| R244A + 1 | 0.017 ± 0.009 | 0.19 ± 0.01 | 11 | ||||
| R244H | 0.022 ± 0.002 | 0.40 ± 0.004 | 18 | ||||
| R244H + 1 | 0.035 ± 0.005 | 0.15 ± 0.007 | 4.3 | ||||
-
HDLs were attached to streptavidin tips via biotinylated lipid, then dipped into LCAT without or with 10 µM compound 1. LCAT was titrated from 0.4 to 2.4 µM, kobs was calculated for each concentration and plotted against concentration. Reactions were performed in triplicate and values are reported as mean ± s.e.m.
Data collection and refinement statistics.
https://doi.org/10.7554/eLife.41604.012| Data collection | ∆N∆C-IDFP·1 Complex (PDB entry 6MVD) |
|---|---|
| Space group | C2 |
| Cell dimensions a, b, c (Å) α, β, γ (°) | 134.5, 106.7, 117.8 90.0, 125.5, 90.0 |
| Resolution (Å) | 30.0–3.10 (3.15–3.10)* |
| Rmerge | 0.115 (≥1) |
| I / σI | 11.1 (1.27) |
| Completeness (%) | 98.9 (100.0) |
| Redundancy | 4.2 (4.2) |
| CC1/2 | (0.55) |
| Refinement | |
| Resolution (Å) | 28.8–3.10 |
| No. reflections | 20,413 |
| Rwork/Rfree | 19.3/23.9 |
| Number of atoms Protein Ligand Water | 6182 5978 183 20 |
| B-factors (Å2) Overall Protein Ligand Water | 73.6 73.2 91.4 41.1 |
| R.m.s. deviations Bond lengths (Å) Bond angles (°) | 0.008 1.33 |
| Ramachandran statistics Favored Allowed Outliers | 93.5 6.0 0.5 |
| MolProbity score | 2.19 |
| Clashscore, all atoms | 4.4 |
-
*Values in parentheses are for the highest-resolution shell.
EC50 values of LCAT variants in the MUP esterase assay with compounds 6, 8, and 9.
https://doi.org/10.7554/eLife.41604.019| EC50 (μM) | |||
|---|---|---|---|
| Variant\Compound | 6 | 8 | 9 |
| WT | no effect | 4.6 ± 0.06 | 7.7 ± 2 |
| Y51S | no effect | >10 | >10 |
| G71I | no effect | >10 | no effect |
| Y51S/G71I | no effect | >10 | no effect |
| R244A | no effect | >10 | 6.2 ± 0.8 |
| R244H | no effect | >10 | 7.6 ± 1 |
-
Compounds were titrated from 0.04 to 9.5 μM, and reactions were performed in triplicate with values reported as mean ± s.e.m.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293F | ThermoFisher | ThermoFisher: R79007 | |
| Cell line (H. sapiens) | HEK293F ∆N∆C-LCAT | this paper | polyclonal stable cell line | |
| Recombinant DNA reagent | pcDNA4 LCAT | (Glukhova et al., 2015) DOI:10.1038/ncomms7250 | ||
| Recombinant DNA reagent | pcDNA4 ∆N∆C-LCAT | (Glukhova et al., 2015) DOI:10.1038/ncomms7250 | ||
| Recombinant DNA reagent | pProEX HT-EndoH | other | D. J. Leahy, Johns Hopkins | |
| Peptide, recombinant protein | ESP24218 peptide | Genscript | PVLDLFRELLNELLEALKQKLK | |
| Commercial assay or kit | Index HT screen | Hampton Research | Hampton: HR2-134 | |
| Commercial assay or kit | Monolith His-Tag Labeling Kit RED-tris-NTA 2nd Generation | Nanotemper Technologies | Nanotemper: MO-L018 | |
| Chemical compound, drug | 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) | Avanti Polar Lipids | Avanti: 850355 | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | NOF America | NOF America: MC-6081 | |
| Chemical compound, drug | 16:0 Biotinyl Cap PE | Avanti Polar Lipids | Avanti: 870277 | |
| Chemical compound, drug | 4-Methylumbelliferyl phosphate | Cayman Chemical | Cayman: 16089 | |
| Chemical compound, drug | Bio-beads SM-2 | Bio-Rad | Bio-Rad: 1523920 | |
| Chemical compound, drug | Bovine Serum Albumin (BSA), fatty acid free | Sigma-Aldrich | Sigma: A8806 | |
| Chemical compound, drug | Cholesterol oxidase | Sigma-Aldrich | Sigma: C8649 | |
| Chemical compound, drug | Dehydroergosterol (DHE) | Sigma-Aldrich | Sigma: E2634 | |
| Chemical compound, drug | DMEM high glucose with GlutaMAX and 1 mM pyruvate | ThermoFisher Scientific | ThermoFisher:10569 | |
| Chemical compound, drug | Fetal bovine serum | Sigma-Aldrich | Sigma: F2442 | |
| Chemical compound, drug | FreeStyle 293 Expression Medium | ThermoFisher Scientific | ThermoFisher: 12338026 | |
| Chemical compound, drug | Isopropyl dodecyl fluorophosphonate | Cayman Chemical | Cayman: 10215 | |
| Chemical compound, drug | Kifunensine | Cayman Chemical | Cayman: 10009437 | |
| Chemical compound, drug | Ni-NTA | Qiagen | Qiagen: 30230 | |
| Chemical compound, drug | OptiMEM Reduced Serum Medium | ThermoFisher Scientific | ThermoFisher:31985070 | |
| Chemical compound, drug | Penicillin- Streptomycin | ThermoFisher Scientific | ThermoFisher: 15140122 | |
| Chemical compound, drug | Polyethylenimine (PEI) | Polysciences | Polysciences: 23966 | |
| Chemical compound, drug | p-nitrophenyl butyrate (pNPB) | Sigma-Aldrich | Sigma: N9876 | |
| Chemical compound, drug | SspI | New England Biolabs | New England Biolabs: R0132S | |
| Chemical compound, drug | SYPRO Orange | ThermoFisher Scientific | ThermoFisher: S6650 | |
| Chemical compound, drug | 6-(4-(4-Hydroxy-6-oxo-4- (trifluoromethyl)−4,5,6,7- tetrahydro-1H-pyrazolo[3,4- b]pyridin-3-yl)piperidin-1-yl) −4- (trifluoromethyl)nicotinonitrile, TFA (compound 1) | example 95 in US patent 9150575 | ||
| Chemical compound, drug | 4-Hydroxy-4-(trifluoromethyl)−3- (1-(5-(trifluoromethyl)pyrazin-2- yl)piperidin-4-yl)−4,5-dihydro-1H- pyrazolo[3,4-b]pyridin-6(7H)-one (compound 2) | example 46 in US patent 9150575 | ||
| Chemical compound, drug | 7-Hydroxy-7-(trifluoromethyl)−1- (1-(5-(trifluoromethyl)pyrazin-2- yl)piperidin-4-yl)−6,7-dihydro-1H- imidazo[4,5-b]pyridin-5(4H)-one (compound 3) | example 3 in WO patent 2015087996A1 | ||
| Chemical compound, drug | 4-(trifluoromethyl)−3-(1-(5- (trifluoromethyl)pyrazin-2- yl)piperidin-4-yl)−1,4,5,7- tetrahydro-6H-pyrazolo[3,4- b]pyridin-6-one (compound 4) | example 10 in WO patent 2015111545A1 | ||
| Chemical compound, drug | 4-Hydroxy-3-(piperidin-4-yl)−4- (trifluoromethyl)−4,5-dihydro-1H- pyrazolo[3,4-b]pyridin-6(7H)-one, HCl (compound 5) | reference example 60 in US patent 9150575 | ||
| Chemical compound, drug | 7-Hydroxy-1-(piperidin-4-yl)−7- (trifluoromethyl)−6,7-dihydro-1H- imidazo[4,5-b]pyridin-5(4H)-one, HCl (compound 6) | reference example 1 in WO patent 2015087996A1 | ||
| Chemical compound, drug | 4-Hydroxy-3-(piperidin-4-yl)−4- (trifluoromethyl)−4,5- dihydroisoxazolo[5,4-b]pyridin- 6(7H)-one, HCl (compound 7) | this paper | Compound was synthesized by NCATS | |
| Chemical compound, drug | 7-(trifluoromethyl)−1-(1-(5- (trifluoromethyl)pyrazin-2- yl)piperidin-4-yl)−1H-imidazo[4,5- b]pyridin-5(4H)-one (compound 8) | this paper | Compound was synthesized by NCATS | |
| Chemical compound, drug | 4-hydroxy-4-(trifluoromethyl)−3- (1-(5-(trifluoromethyl)pyrazin-2- yl)piperidin-4-yl)−4,5- dihydroisoxazolo[5,4-b]pyridin- 6(7H)-one (compound 9) | this paper | Compound was synthesized by NCATS | |
| Software, algorithm | HKL-2000 | (Otwinowski and Minor, 1997) DOI:10.1016/S0076-6879(97)76066-X | ||
| Software, algorithm | PHASER | (McCoy, 2007) DOI:10.1107/ S0907444906045975 | ||
| Software, algorithm | REFMAC5 | (Murshudov et al., 2011) DOI:10.1107/ S0907444911001314 | ||
| Software, algorithm | Phenix | (Adams et al., 2010) DOI:10.1107/ S0907444909052925 | ||
| Software, algorithm | Coot | (Emsley et al., 2010) DOI:10.1107/S090 7444910007493 | ||
| Software, algorithm | Molprobity | (Chen et al., 2010) DOI:10.1107/S0907 444909042073 | ||
| Software, algorithm | Prism 7.0 c | Graphpad Software | ||
| Software, algorithm | Octet Data Analysis 7.0 | FortéBio | ||
| Software, algorithm | PyMOL Molecular Graphics System | Schrödinger | ||
| Software, algorithm | Chimera | (Pettersen et al., 2004) DOI:10.1002/jcc.20084 | ||
| Software, algorithm | Protein Thermal Shift | ThermoFisher Scientific | ||
| Software, algorithm | MO.Affinity Analysis | Nanotemper Technologies | ||
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.41604.027





