FOXP2 exhibits projection neuron class specific expression, but is not required for multiple aspects of cortical histogenesis
Figures
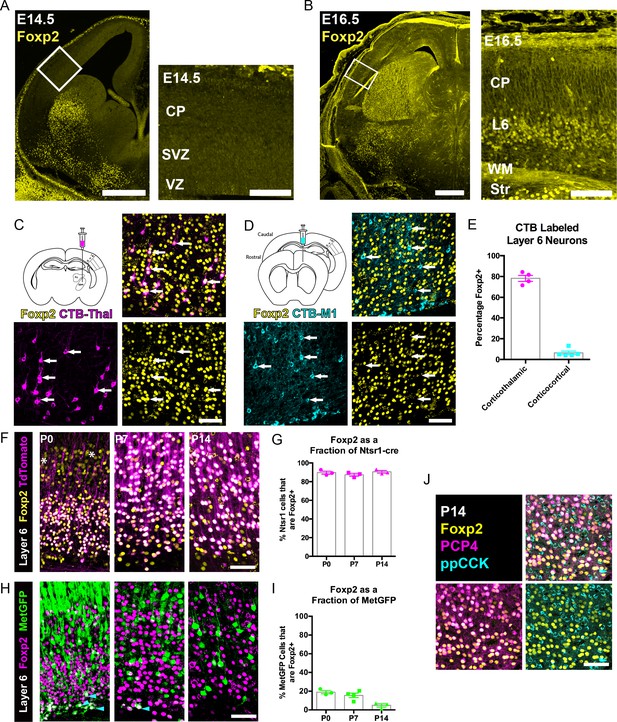
FOXP2 is enriched in corticothalamic neurons during cortical development.
(A) Low magnification (left) and high magnification (right) images of FOXP2 (yellow) immunohistochemical labeling of E14.5 forebrain reveals absence of expression in the dorsal pallium, whereas the developing striatum is robustly labeled at this timepoint (N = 5). (B) Images of FOXP2 immunolabeling at E16.5 demonstrates the presence of FOXP2 expression within the deep layers of the developing cortical plate (N = 3). (C) Retrograde labeling of layer 6 corticothalamic neurons (magenta) by injection of CTB into the ventrobasal thalamus, combined with FOXP2 (yellow) immunohistochemistry at P14. (D) Corticocortical neurons (cyan) labeled by injection of CTB into the ipsilateral primary motor cortex, combined with FOXP2 (yellow) immunohistochemistry at P14. White arrows denote retrogradely labeled projection neurons. (E) Quantification of the percentages of retrogradely labeled corticothalamic (N = 4 mice) and corticocortical (N = 5) neurons that express FOXP2. (F) FOXP2 (yellow) immunohistochemistry in sections of P0, P7, and P14 somatosensory cortex from Ntsr1-cre; tdTomato mice (tdTomato is magenta); white asterisks denote relatively low-level expression in layer 5 at P0. (G) Quantification of the percentages of tdTomato-positive neurons that express FOXP2 at each age (P0, N = 3; P7, N = 3; P14, N = 3). (H) FOXP2 (magenta) immunohistochemistry in sections of P0, P7, and P14 somatosensory cortex from MetGFP (green) mice. Cyan arrowheads denote sparse FOXP2+ and GFP+ double-labeled cells localized to layer 6B/subplate. (I) Quantification of the percentages of GFP+ neurons that co-express Foxp2 at each age (P0, N = 3; P7, N = 4; P14, N = 3). (J) FOXP2 (yellow) colocalizes with PCP4 (magenta), but not ppCCK (cyan) (N = 3). Scale bars: 500 µm, A, B low magnification; 100 µm A, B high magnification; 50 µm C, D, F, H and J.
-
Figure 1—source data 1
Foxp2 expression among retrogradely labeled and molecularly defined developing layer 6 neuron classes.
- https://doi.org/10.7554/eLife.42012.005
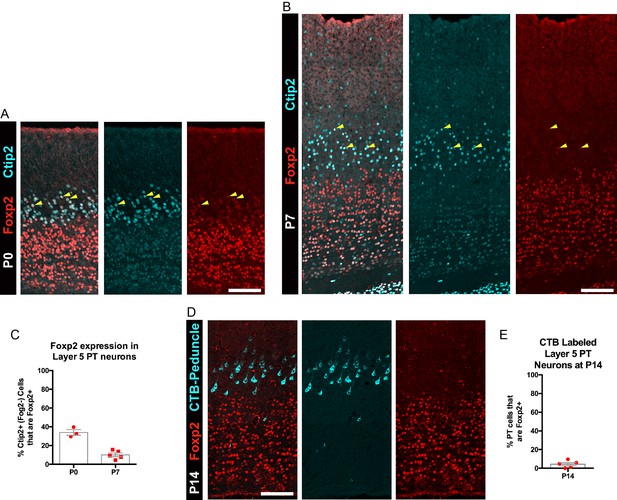
FOXP2 is transiently expressed by a subpopulation of PT neurons.
(A) FOXP2 (red) and CTIP2 (cyan) immunohistochemistry in sections of P0 somatosensory cortex. (B) FOXP2 (red) and CTIP2 (cyan) immunohistochemistry in sections of P7 somatosensory cortex. (C) Quantification of the percentage of CTIP2+ PT neurons in somatosensory cortex that express FOXP2 at P0 (N = 3) and P7 (N = 5). (D) Lack of expression of FOXP2 in most retrogradely (CTB) labeled PT neurons at P14 (N = 5). (E) Quantification of the percentage of CTB-labeled PT neurons that express FOXP2. All scale bars, 100 µm.
-
Figure 1—figure supplement 1—source data 1
Foxp2 expression among retrogradely and molecularly defined developing pyramidal tract neurons.
- https://doi.org/10.7554/eLife.42012.004
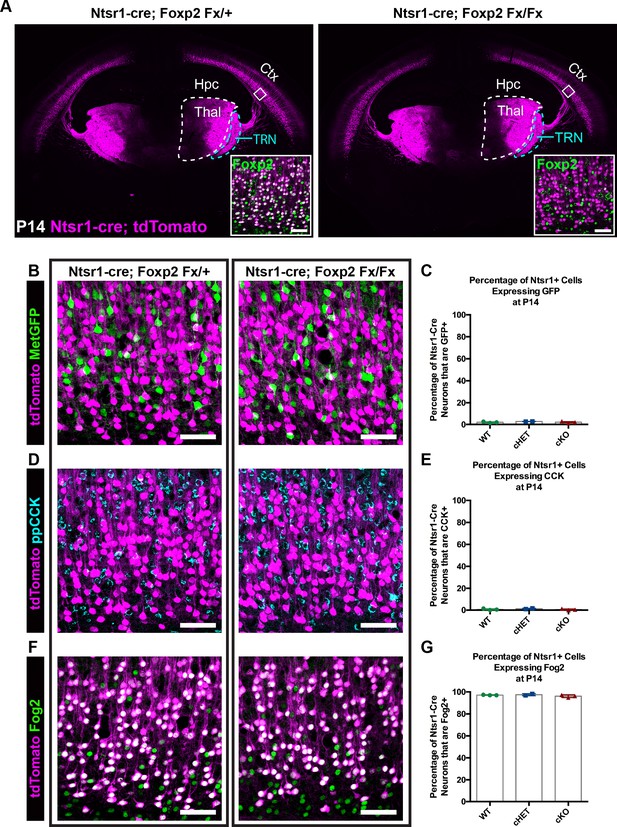
FOXP2 is nonessential for class-specific anatomical and molecular phenotypes of corticothalamic neurons.
(A) At P14 tdTomato (magenta) expression in layer 6 corticothalamic neurons of Ntsr1-cre; Rosa-tdTomato mice reveals similar organization of corticothalamic innervation in Foxp2 conditional knockout mice and heterozygous littermates – boxed inset shows removal of FOXP2 protein (green) from tdTomato+ corticothalamic neurons of Ntsr1-cre; Foxp2Fx/Fx mouse. (B) MetGFP (green) and tdTomato (magenta) label distinct cell populations in Foxp2 conditional knockout mice, heterozygous littermates, and wild-type C57Bl/6J mice. (C) Quantification of co-expression of GFP by tdTomato+ corticothalamic neurons across Foxp2 genotypes (WT = Ntsr1 cre; Rosa-tdTomato, no Flox alleles, N = 3 mice; cHET = Ntsr1 cre; Foxp2Fx/+, N = 2 mice; cKO = Foxp2 Fx/Fx, N = 3 mice). (D) ppCCK expression is excluded from layer 6 corticothalamic neurons across Foxp2 genotypes as indicated by the segregation of tdTomato (magenta) and ppCCK (cyan). (E) Quantification of co-expression of CCK by tdTomato+ corticothalamic neurons (N for each group same as panel C). (F) FOG2 expression by corticothalamic neurons does not require Foxp2, as nearly all tdTomato+ cells express FOG2 (green) across Foxp2 genotypes. (G) Quantification of FOG2 coexpression by tdTomato+ corticothalamic neurons (N for each group same as panel C). All scale bars, 50 µm. Abbreviations: Ctx, cortex; Hpc, hippocampus; Thal, thalamus; TRN, thalamic reticular nucleus.
-
Figure 2—source data 1
Expression of Layer 6 cell-type markers in Ntsr1-cre; Foxp2Fx mice and controls.
- https://doi.org/10.7554/eLife.42012.008
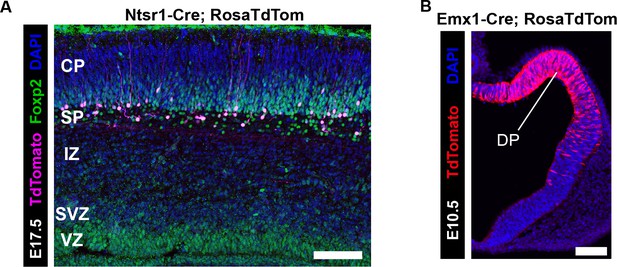
Developmental timing of Cre-mediated recombination in Ntsr1-cre and Emx1-cre.
(A) Cre-dependent tdTomato (magenta) expression begins at E17.5 (N = 3) in the cortex of Ntsr1-cre mice, when there are abundant FOXP2+ (green) neurons in the subplate and layer 6 that do not yet express tdTomato. (B) Cre-dependent tdTomato (red) expression begins at E10.5 (N = 3) in the cortex of Emx1-cre mice, where the majority of dorsal pallium (DP) progenitors are tdTomato+. (CP, cortical plate; IZ, intermediate zone; Str, striatum; SVZ, subventricular zone; VZ, ventricular zone; WM, white matter). Scale Bars: 100 µm.
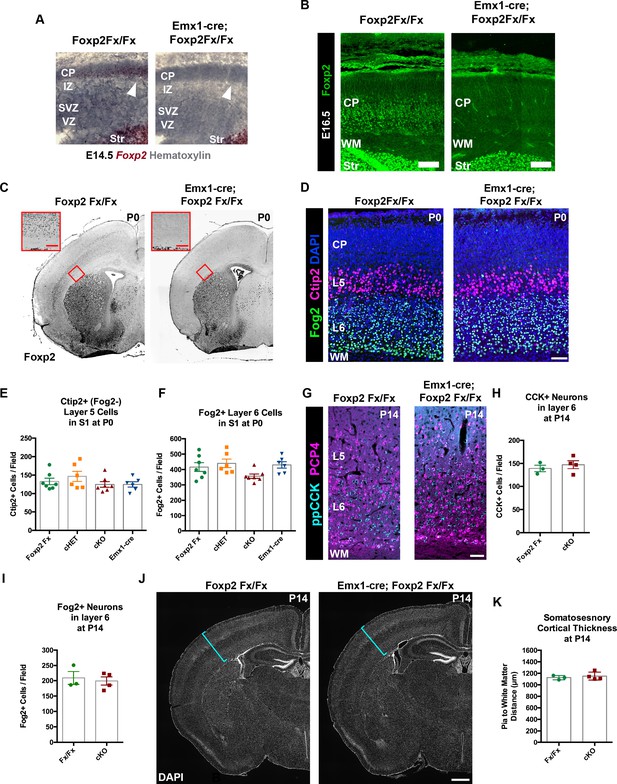
FOXP2 is nonessential for the genesis of cortical neurons and their proper lamination.
(A) Foxp2 in situ hybridization based on the BaseScope method reveals expression of Foxp2 transcript (Red) in Foxp2Fx/Fx embryos (N = 4) and selective removal of exons 12–14 (DNA-binding domain) from the dorsal pallium including the cortical plate (white arrowhead) of Emx1-cre; Foxp2Fx/Fx mice (N = 6) by E14.5. (B) Immunohistochemical analysis of FOXP2 protein in E16.5 embryos (N = 3 each genotype) demonstrates selective elimination of FOXP2 protein (green) from the infragranular layers of the dorsal pallium of Emx1-cre;Foxp2Fx/Fx mouse embryos. (C) FOXP2 immunohistochemistry on coronal sections of P0 Foxp2Fx/Fx and Emx1-cre; Foxp2Fx/Fx mice reveals absence of FOXP2 (black) in the infragranular cortical layers (red bracket) of Emx1-cre; Foxp2Fx/Fx mice.Inset (red outline) shows selective loss of FOXP2 in the cortex at higher magnification. (D) FOG2 (green) and CTIP2 (magenta) immunohistochemistry in coronal sections of the primary somatosensory cortex of Foxp2Fx/Fx and Emx1-cre; Foxp2Fx/Fx mice reveals similar distributions of laminar specific markers at P0. (E) Quantification of FOG2+ cells in layer 6 across genotypes at P0 (Fx/Fx, Foxp2fx/fx, N = 7; cHET, Emx1-cre; Foxp2fx/+, N = 6; cKO, Emx1-cre;Foxp2fx/fx, N = 7; Emx1-cre, N = 6). (F) Quantification of CTIP2+/Fog2- cells in layer 5 across genotypes at P0 (N for each group, same as panel E). (G) ppCCK (cyan) and PCP4 (magenta) immunohistochemistry in coronal sections of conditional knockout and control littermates at P14. (H) Quantification of ppCCK+ cells in layer 6 of SSC across genotypes at P14 (Foxp2Fx/Fx, N = 3 mice; Emx1-cre; Foxp2Fx/Fx, N = 4 mice). (I) Quantification of FOG2+ cells in layer 6 of SSC across genotypes at P14 (Foxp2Fx/Fx, N = 3 mice; Emx1-cre; Foxp2Fx/Fx, N = 4 mice). (J) DAPI-staining of coronal sections of Foxp2Fx/Fx and Emx1-cre; Foxp2Fx/Fx mice reveals similar size of cortex, including the thickness of primary somatosensory cortex (indicated by cyan bracket). (K) Quantification of somatosensory cortex thickness across genotypes at P14 (Foxp2Fx/Fx, N = 3 mice; Emx1-cre; Foxp2Fx/Fx, N = 4 mice). Scale Bars: A(inset), 100 µm; B, E, 50 µm; H, 500 µm.
-
Figure 3—source data 1
Quantification of cortical cell type numbers in Emx1-cre; Foxp2Fx mice and controls.
- https://doi.org/10.7554/eLife.42012.011
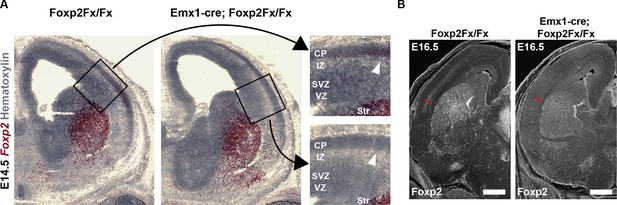
Elimination of Foxp2 transcript and protein from the forebrain of Emx1-cre; Foxp2Fx/Fx embryos.
(A) BaseScope RNAscope demonstrating selective removal of Foxp2 exons 12–14 (DNA-binding domain) from the dorsal pallium at E14.5 (note that high magnification insets are the same images as Figure 3A). (B) FOXP2 immunohistochemistry demonstrates selective removal of FOXP2 protein from the infragranular layers (red bracket) of the dorsal pallium at the earliest timepoint that it can be detected, E16.5. Scale bar = 500 µm.
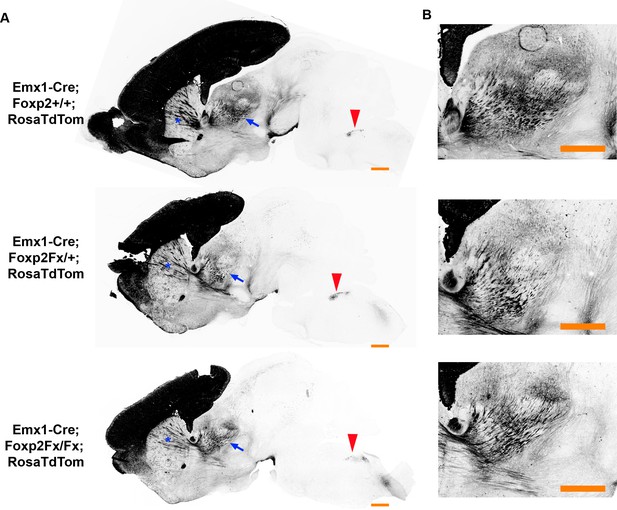
FOXP2 is not required for proper corticofugal axon pathfinding.
(A) tdTomato reporter (black) reveals similar patterns of corticofugal axon growth in sagittal sections of Emx1-cre;Foxp2+/+ (WT, top panel, N = 4) Emx1-cre; Foxp2Fx/+ (middle, N = 3) and Emx1-cre; Foxp2Fx/Fx (bottom, N = 3). Note the fasciculation of axons in the internal capsule (blue asterisks) and the comparable growth of axons into the thalamus (blue arrows) and pyramidal decussation (red arrowheads). (B) Higher magnification images of the corticothalamic innervation patterns in each genotype. Scale Bars = 500 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Foxp2Fx (Foxp2tm1.1Sfis) | French et al., 2007 | MGI Cat# 3800702, RRID:MGI:3800702 | |
| Genetic reagent (M. musculus) | Rosa-TdTomato (Ai14) | Jackson Laboratory | IMSR Cat# JAX:007914, RRID:IMSR_JAX:007914 | |
| Genetic reagent (M. musculus) | Ntsr1-cre (GN220) | MMRRC | MMRRC Cat# 030648-UCD, RRID:MMRRC_030648-UCD | |
| Genetic reagent (M. musculus) | Emx1-cre | Gorski et al., 2002 | IMSR Cat# JAX:005628, RRID:IMSR_JAX:005628 | |
| Genetic reagent (M. musculus) | MetEGFP BAC (MetGFP) | Gong et al., 2003 | MGI:6144427 | |
| Antibody | Goat anti-Foxp2 (polyclonal) | Santa Cruz Biotechnology | Cat# sc-21069, RRID:AB_2107124 | IHC (1:100) |
| Antibody | Chicken anti-GFP (polyclonal) | Abcam | Cat# ab13970, RRID:AB_300798 | IHC (1:500) |
| Antibody | Rat anti-Ctip2 (monoclonal) | Abcam | Cat# ab18465, RRID:AB_2064130 | IHC (1:500) |
| Antibody | Guinea Pig anti-ppCCK | Watakabe et al., 2012 | Dr. Takeshi Kaneko (University of Tokyo) | IHC (1:500) |
| Antibody | Rabbit anti-PCP4(PEP-19) | Dr. James Morgan (St. Jude's Research Hospital) | IHC (1:3000) | |
| Antibody | Rabbit anti-Fog2 (polyclonal) | Santa Cruz Biotechnology | Cat# sc-10755, RRID:AB_2218978 | IHC (1:250) |
| Antibody | Rabbit anti-DARPP-32 (monoclonal) | Cell Signaling Technology | Cat# 2306, RRID:AB_823479 | IHC (1:500) |
| Antibody | AlexaFluor F(AB')2 488- or 594- or 647- secondaries | Jackson Immunoresearch Laboratories, Inc | IHC (1:500) | |
| Peptide, recombinant protein | Cholera Toxin Subunit B (CTB) | Invitrogen | Cat. #: C-34776, C-34778 | Alexa Fluor Conjugate (555, 647) |
| Commercial assay or kit | BaseScope assay | Advanced Cell Diagnostics | Cat. #: 323971 | |
| Software, algorithm | IMARIS | Imaris (http://www.bitplane.com/imaris/imaris) | RRID:SCR_007370 | Microscopy Image Analysis Software |
| Software, algorithm | Adobe Photoshop | Adobe Photoshop (https://www.adobe.com/products/photoshop.html) | RRID:SCR_014199 | |
| Software, algorithm | Adobe Illustrator (CS6) | Adobe Illustrator (http://www.adobe.com/products/illustrator.html) | RRID:SCR_010279 | |
| Software, algorithm | Code used for nuclear immunofluorescent quantification | This paper | custom written for ImageJ macros (Source Code 1; Source Code 2) | |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism 6 | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 6 |
Additional files
-
Source code 1
Maximum Projection and Cropping Macro.
- https://doi.org/10.7554/eLife.42012.013
-
Source code 2
Cortical Cell Type Autocounting Macro for nuclear markers.
- https://doi.org/10.7554/eLife.42012.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42012.015





