Screening for insulin-independent pathways that modulate glucose homeostasis identifies androgen receptor antagonists
Figures
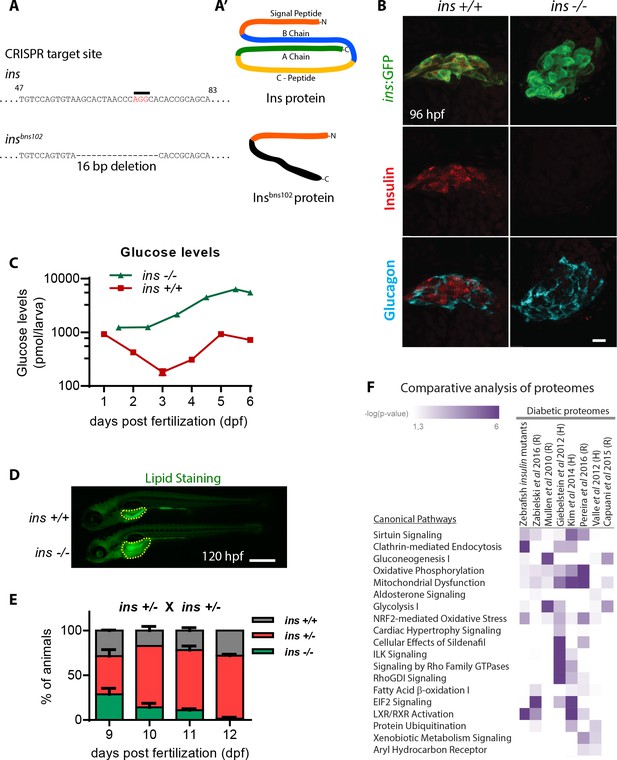
insulin is crucial for zebrafish metabolic homeostasis.
(A) CRISPR target site in the insulin gene, with PAM sequence highlighted, and the resulting 16 bp deletion allele (below). (A’) Schematic of wild-type Insulin protein and the predicted mutant protein which contains novel sequence (black). (B) Confocal projection images of the pancreatic islet in 96 hpf Tg(ins:GFP) ins +/+ and ins -/- animals immunostained for Insulin (red) and Glucagon (cyan). (C) Free glucose levels in wild-type and mutant animals from 1 to 6 dpf; mean ± SEM, n = 2–4 replicates. (D) Nile Red staining (green) for neutral lipids in 120 hpf wild-type (top) and mutant (bottom) larvae, with yolk lipid content outlined (yellow dots). (E) Genotype distribution from ins ± incross, calculated as the percentage of total animals at each stage; mean ± SEM, n = 32 animals at each stage. (F) Heat map of the proteomic signature of zebrafish ins mutants at 120 hpf compared to signatures from rodent (R) and human (H) diabetic proteome studies. Canonical pathways implicated in most studies are listed first. P-value cut-off set at <0.05. Scale bars: 10 μm (B), 500 μm (D).
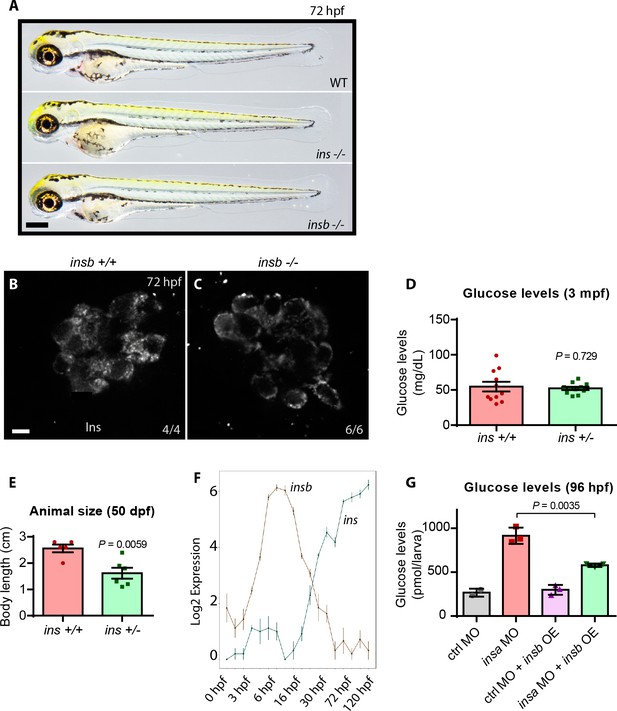
ins, and not insb, is the predominant paralog expressed in zebrafish pancreatic islets.
(A) Brightfield images of 72 hpf wild-type, ins mutant and insb mutant larvae. (B–C) Confocal plane images of the pancreatic islet in 72 hpf wild-type and insb mutant larvae stained for Insulin (white). (D) Fasting blood glucose levels in three mpf wild-type and ins ± animals; mean ± SEM, n = 11 animals. (E) Body length measurements of 50 dpf wild-type and ins ± animals; mean ± SEM, n = 5–6 animals. (F) mRNA expression time course of ins and insb during zebrafish development from 0 to 120 hpf. (G) 4 ng of control (ctrl) or ins morpholino (MO) was injected in non-transgenic or Tg(ins:insb) (insb OE) embryos at the one cell stage and glucose levels measured at 96 hpf; mean ± SEM, n = 3 replicates. Scale bars: 250 μm (A), 5 μm (C, D). P values from t-tests.
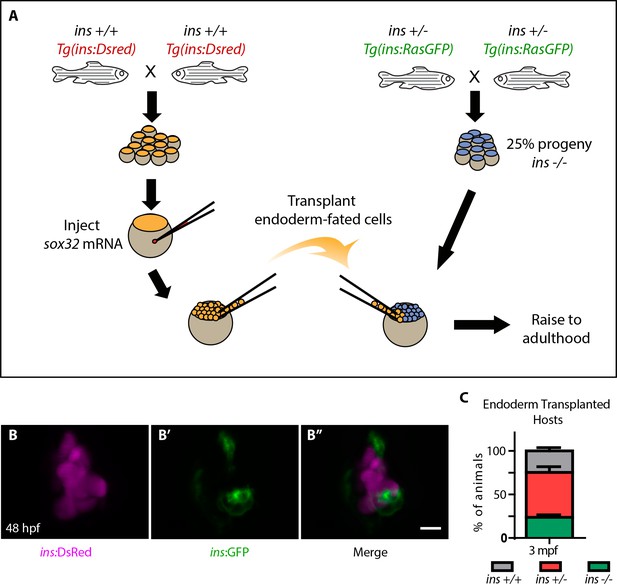
Highly efficient endoderm transplant technique rescues ins mutants to adulthood.
(A) Schematic depicting the endoderm transplantation protocol; sox32 mRNA-injected ins +/+ donor cells (orange) were transplanted into host embryos (blue) to form chimeric animals. (B–B’’) Confocal projection images of the pancreatic islet of a 48 hpf chimeric animal showing β-cells from the host (green, (B’) and the transplanted ins +/+ cells (magenta, (B). (C) Genotype distribution in the raised three mpf chimeric animals, determined by genotyping fin tissue; mean ± SEM, n = 3 transplant experiments, 18–32 animals per experiment. Scale bar: 10 μm.
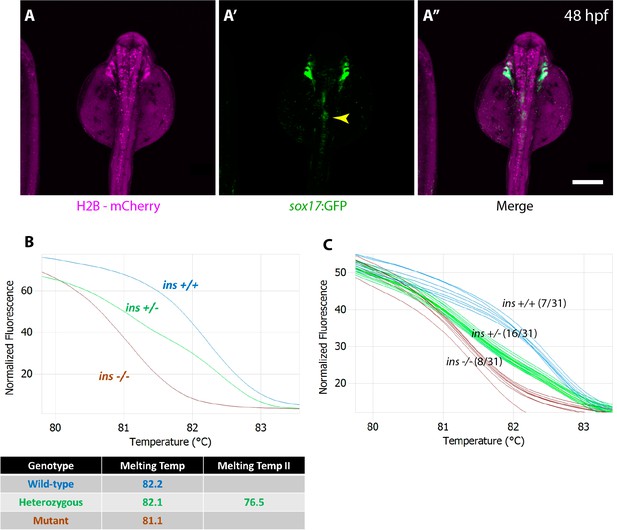
sox32 mRNA-injected cells contribute to host endoderm upon transplantation.
(A–A”) Confocal images of a 48 hpf host embryo injected with H2B-mCherry mRNA (magenta) and transplanted with Tg(sox17:GFP) expressing donor endoderm showing the chimeric islet (yellow arrowhead). (B) High-resolution melt analysis patterns for genotyping ins +/+ (blue), ins +/- (green) and ins -/- (brown) animals. (C) Representative example of genotyping 31 chimeric adults, revealing 8 of 31 animals to be mutant (brown). Scale bar: 200 μm.
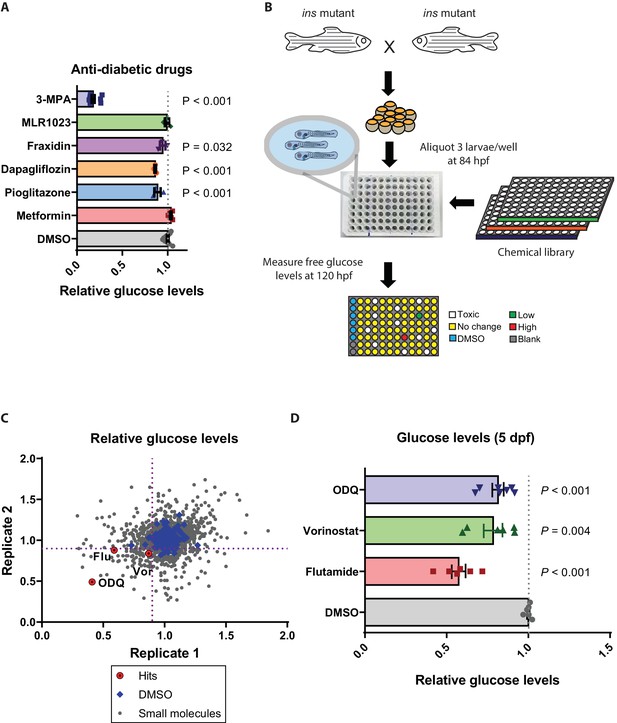
Small molecule screen in ins mutants reveals insulin-independent modulators of glucose metabolism.
(A) Relative glucose levels in 120 hpf ins mutant larvae after 36 hr of treatment with anti-diabetic drugs (dapagliflozin, pioglitazone, metformin) or reported insulin mimetics (MLR1023, Fraxidin) or the Pck1 inhibitor, 3 MPA; mean ± SEM, n = 3–7 replicates. (B) Schematic representation of the screening pipeline: ins mutant larvae were treated with small molecules starting at 84 hpf and free glucose levels measured at 120 hpf. (C) Scatter plot showing relative change in glucose levels upon treatment with 2233 small molecules. X and Y axes represent two replicates performed for each drug, with the dotted purple lines marking 0.9 on each axis. 72 molecules satisfied the pre-specified cut-off. (D) Relative glucose levels at 120 hpf upon treatment of ins mutants with the three hits – ODQ, Vorinostat, and Flutamide; mean ± SEM, n = 6–7 replicates. P values from t-tests.
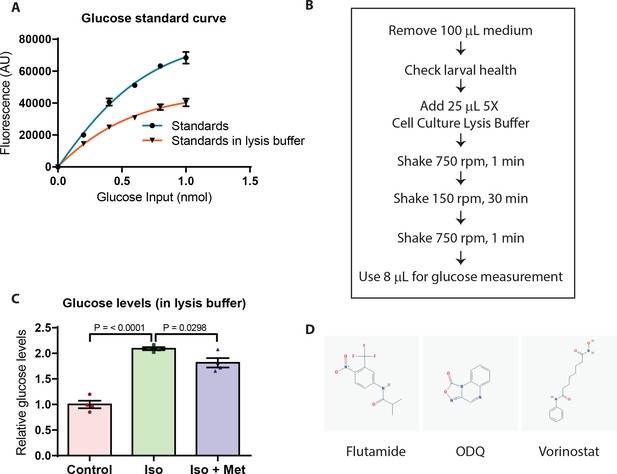
A 96-well plate-adapted protocol to measure glucose levels is suitable for small molecule screening.
(A) Plotting the measured glucose levels interpolated from the regular standard curve (blue) shows a shifted curve in the presence of lysis buffer (orange) with a dynamic range still suitable for measuring glucose levels. (B) Schematic showing the plate-adapted glucose readout assay. (C) Relative free glucose levels upon treatment of wild-type larvae from 4 to 6 dpf with the beta-adrenergic agonist isoprenaline (Iso) or both isoprenaline and metformin (Iso +Met); mean ± SEM, n = 4–5 replicates. (D) Structures of the hits - flutamide, ODQ and vorinostat. P values from t-tests.
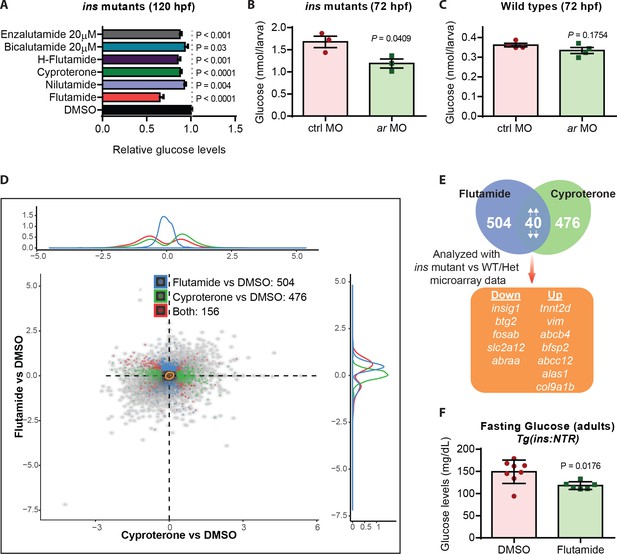
Androgen receptor (AR) antagonism reduces glucose levels in hyperglycemic larvae and adults.
(A) Relative glucose levels in ins mutants at 120 hpf upon treatment with various AR antagonists starting at 84 hpf; mean ± SEM, n = 3–7 replicates. (B) Glucose levels in 72 hpf ins mutants after injection with 1 ng of ctrl or ar MO; mean ± SEM, n = 3 replicates. (C) Glucose levels in 72 hpf wild types after injection with 1 ng of ctrl or ar MO; mean ± SEM, n = 4 replicates. (D) RNA-seq analysis of 120 hpf ins mutant larvae treated with flutamide or cyproterone starting at 84 hpf, showing differentially expressed genes (DEGs) compared to DMSO-treated larvae in blue and green, respectively. Red dots indicate DEGs common to both treatments. (E) Workflow used to filter candidate genes: 40 DEGs modulated in the same direction (both up or both down) were analyzed in relation to the microarray dataset (ins mutant vs phenotypically wild-type 108 hpf larvae). (F) Glucose levels measured in adult Tg(ins:NTR) animals after β-cell ablation and intraperitoneal injection with vehicle (DMSO) or flutamide; mean ± SEM, n = 6–8 animals. P values from t-tests.
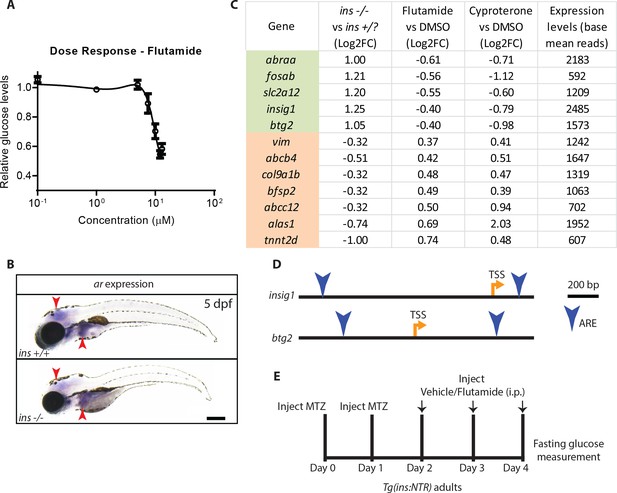
Flutamide reduces glucose levels in a dose-dependent manner, possibly exerting its effects through liver gluconeogenic enzymes.
(A) Dose response curve of the glucose lowering effect of flutamide in ins mutant larvae, treated from 84 to 120 hpf; mean ± SEM, n = 3 replicates. (B) Wholemount in situ hybridization for ar expression in 120 hpf wild-type and ins mutant larvae showing signal in the brain and liver (red arrowheads). (C) The 12 genes differentially regulated in ins mutants compared to non-mutant siblings and modulated in the opposite direction upon treatment with flutamide or cyproterone compared to DMSO; listed with their fold change (Log2 scale) and expression levels under control condition (base mean reads); genes are ordered by the fold change in the flutamide vs DMSO condition. (D) Schematic of insig1 and btg2 gene loci, with location of transcription start site (TSS) and androgen response elements (ARE, blue arrowheads). (E) Schematic of vehicle vs flutamide treatment of Tg(ins:NTR) adult animals following β-cell ablation with metronidazole (MTZ) injection to induce hyperglycemia. Scale bar: 250 μm.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Danio rerio) | insbns102 | This paper | 16 bp deletion allele of ins (Gene ID: 30262) | |
| Antibody | α-Insulin (Guinea Pig Polyclonal) | Dako | A0564 | (1:300) |
| Sequence-based reagent | sox32 | Kikuchi et al., 2001 | (RNA) | |
| Commercial assay or kit | mMessage mMachine SP6 Transcription Kit | ThermoFisher | AM1340 | |
| Commercial assay or kit | Glucose assay kit | Merck | CBA086 | |
| Software, algorithm | ZEN Blue 2012 | Zeiss, Germany | ||
| Software, algorithm | GraphPad Prism 7 | GraphPad Software, California |
Additional files
-
Supplementary file 1
List of proteins with Log2FC > 1 or Log2FC < −1 from proteomic analyses comparing 120 hpf ins mutant and wild-type animals.
- https://doi.org/10.7554/eLife.42209.011
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42209.012





