Symbiont location, host fitness, and possible coadaptation in a symbiosis between social amoebae and bacteria
Figures
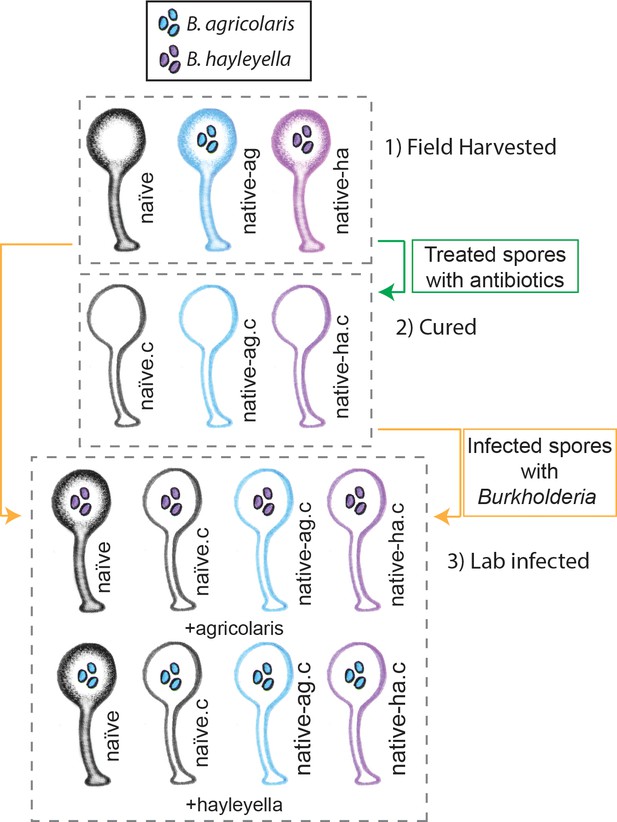
Illustration of host-symbiont pairs used throughout the study.
D. discoideum clones were originally harvested from the wild in three different states: uninfected (indicated as naïve), or naturally infected with B. agricolaris or B. hayleyalla (indicated as native-ag, and native-ha respectively). Clones were treated with antibiotics to eliminate symbionts and are indicated with a ‘.c’. Clones were subsequently exposed to Burkholderia to initiate new infections. Thus, experimental types include 1) Field harvested, 2) cured, and 3) lab infected hosts.
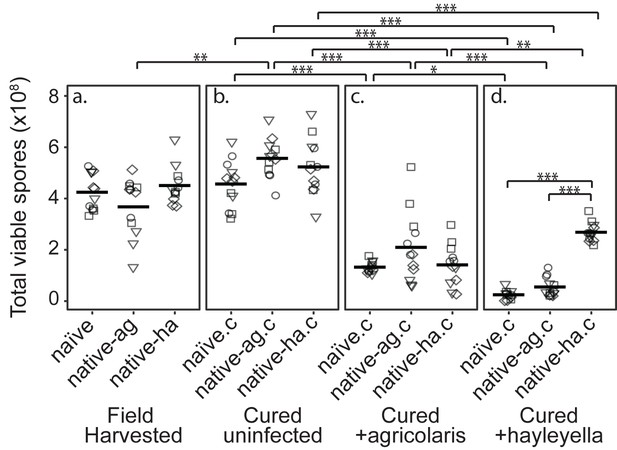
Burkholderia Infections Differentially Alter Spore Viability According to Burkholderia Species and Host Background.
Total viable spores were determined for naïve and native hosts in their field harvested (a), cured (b), B. agricolaris lab-infected (c), and B. hayleyella lab-infected state (d). Four clones were measured for each type with three replicates for each (squares, triangles, circles, and diamonds represent set 1–4 clones respectively). Spore viability for wild harvested B. agricolaris and B. hayleyella host clones is higher than their cured-re-infected counterparts. Notably, spores from infected B. agricolaris and B. hayleyella native hosts (either naturally infected or cured and re-infected with their original Burkholderia) have a higher fitness than Burkholderia infected non-native counterparts. Bars represent significant differences (p < 0.05, and as indicated in supplemental tables).
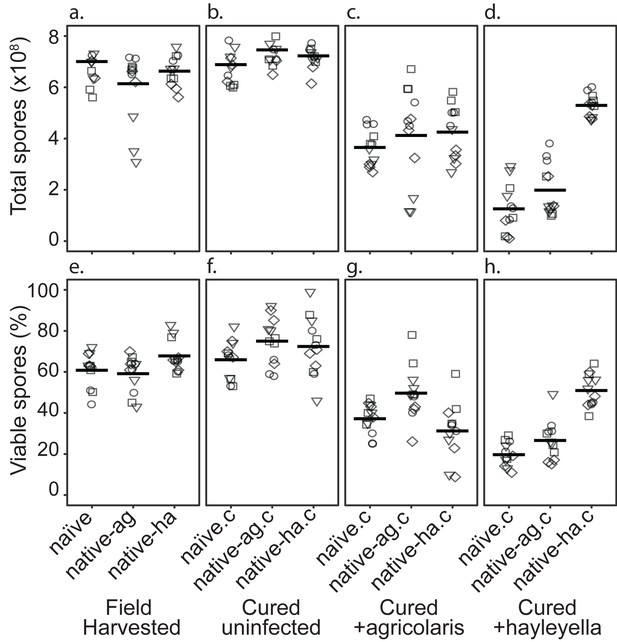
Total Spore Number and Percent of Viable Spores for Burkholderia Infections in Diverse Host Backgrounds.
Total spores (top panel) and percent viable spores (bottom panel) were determined for naïve and native hosts in their field harvested (a), cured (b), B. agricolaris lab-infected (c), and B. hayleyella lab-infected state (d). Four clones were measured for each type with three replicates for each (squares, triangles, circles, and diamonds represent set 1–4 clones respectively). These data were used to determine total viable spores represented in Figure 2.
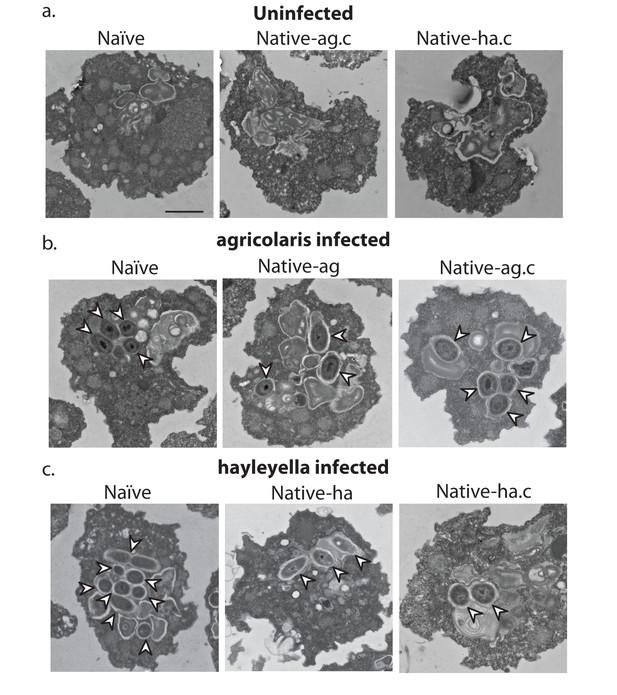
Bacterial cells are found within Burkholderia exposed vegetative amoebae.
Transmission electron micrographs of vegetative amoebae show naïve and cured native amoebae with intracellular morphologies suggestive of active bacterial digestion with no evidence of intact intracellular bacteria (a). In contrast, bacterial cells can be found within B. agricolaris (b) and B. hayleyella (c) infected hosts. Arrows point to bacterial cells. More bacteria are observed in the B. hayleyella infected naïve host than in field harvested native-hayleyella and cured and re-infected native-hayleyella hosts (c). Bacterial cells appear to be within vacuole-like compartments. Scale bar (applicable to all): 2 um.
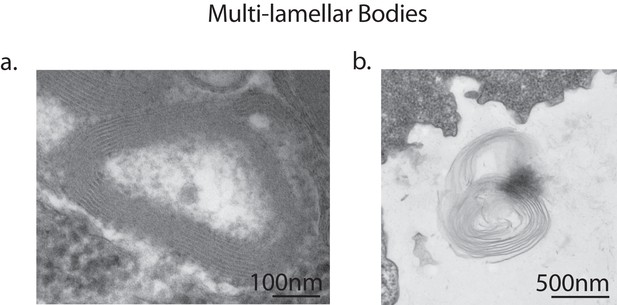
Multi-lamellar bodies excreted by vegetative amoebae.
Transmission electron micrographs of vegetative amoebae identified multi-lamellar bodies inside uninfected amoebae, indicating successful digestion of bacterial food (a). Multi-lamellar bodies are eventually secreted into the surrounding medium (b).
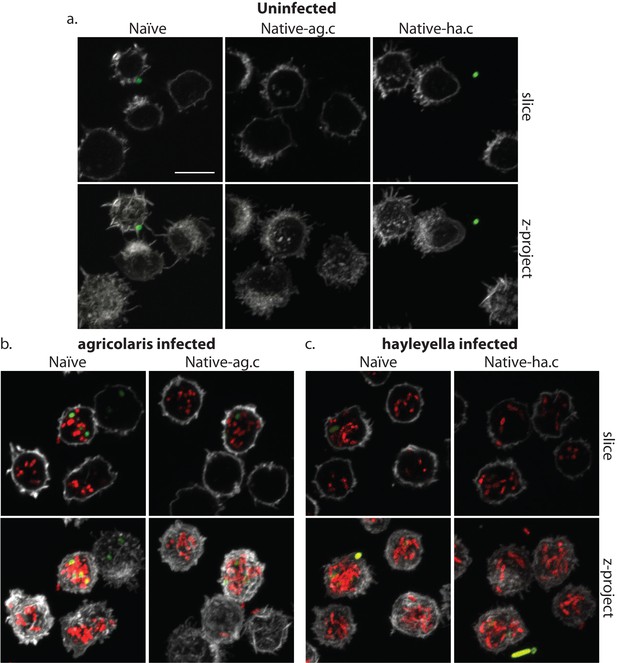
Burkholderia is found abundantly in colonized vegetative amoebae.
Confocal imaging of fixed and stained vegetative amoebas show little to no intracellular bacteria in uninfected clones (a). However, abundant Burkholderia (Burkholderia-RFP shown in red) is found in B. agricolaris (b) and B. hayleyella (c) infected hosts. Occasional intracellular food bacteria (Klebsiella-GFP shown in green) is seen in B. agricolaris hosts (c). Spore coats are stained with phalloidin shown in grey. Scale bar 10 um.
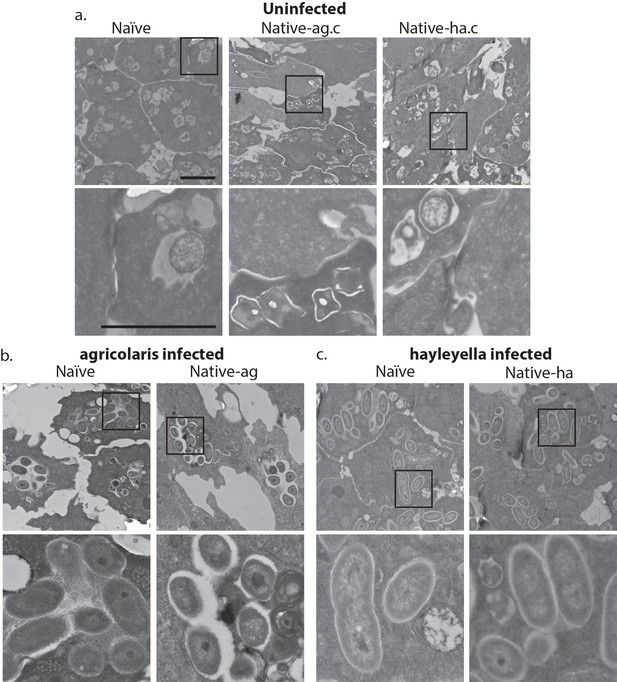
Intracellular bacteria are retained in naïve migrating slugs exposed to Burkholderia and in native Burkholderia hosts.
Transmission electron micrographs of uninfected (a) show closely packed amoebae with internal structures reminiscent of previous bacterial digestion but without evidence of intact internal bacteria. In contrast, B. agricolaris (b) and B. hayleyella (c) infected slugs retain intracellular bacteria. Bottom panels represent magnified versions (see box) of upper panels. Scale bar (applicable to all panels in row) 2 um.
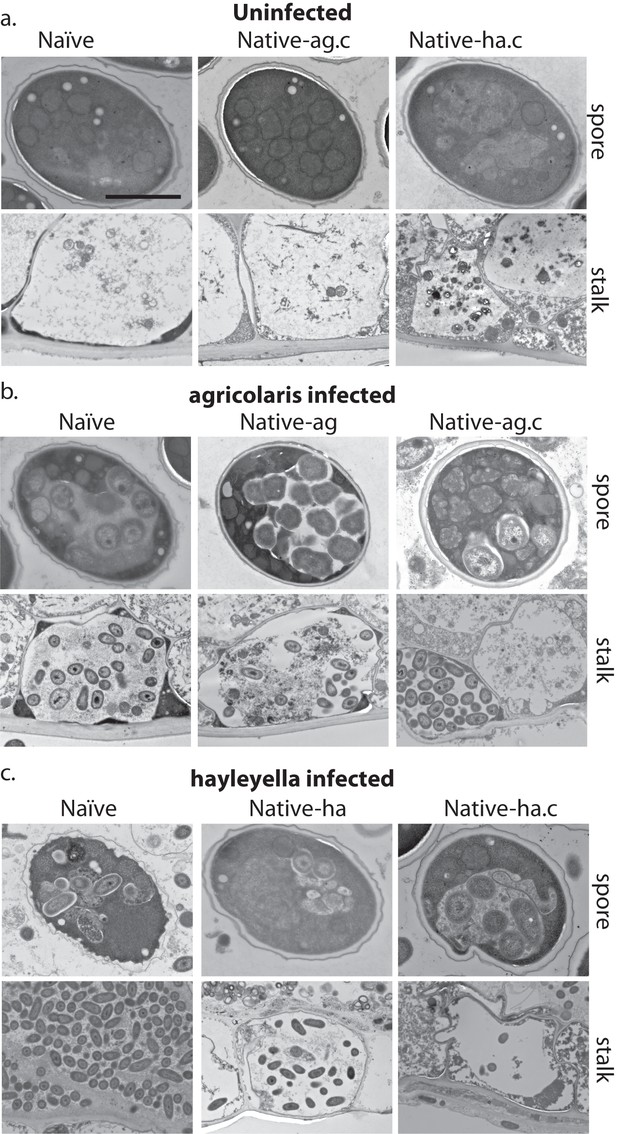
Bacterial cells are retained in spore and stalk cells from Burkholderia-exposed hosts.
As visualized through transmission electron microscopy, (a) uninfected hosts form sturdy spores and stalk cells with no detectable bacteria. Spores and stalk cells retain intracellular bacteria in B. agricolaris (b) and B. hayleyella (c) hosts. Naïve B. agricolaris hosts appear structurally similar to uninfected cells while naïve B. hayleyella hosts have compromised spore coats and collapsed stalk structures filled with bacteria. Scale bar: 2 um.
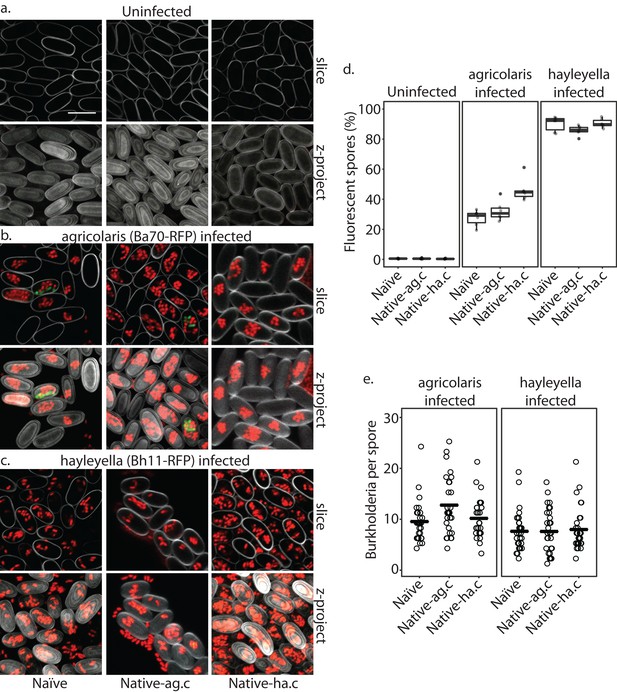
Burkholderia is retained in the sori of developed D. discoideum hosts and the percent of Burkholderia positive spores differs according to Burkholderia species.
Confocal images show no intra- or extracellular bacteria in uninfected spores (a) Abundant Burkholderia is seen in B. agricolaris (b) and B. hayleyella (c) hosts, with more infected spores seen for B. hayleyella (d) but more Burkholderia-RFP cells detected per infected spore for B. agricolaris hosts (e). Co-infection by food bacteria is occasionally observed in B. agricolaris infected spores (b). For a-c: Klebsiella-GFP shown in green, Burkholderia-RFP shown in red, and calcofluor stain shown in grey. Top panels are image slices; bottom panels are max intensity projections of z stacks. Scale bar: 10 um.
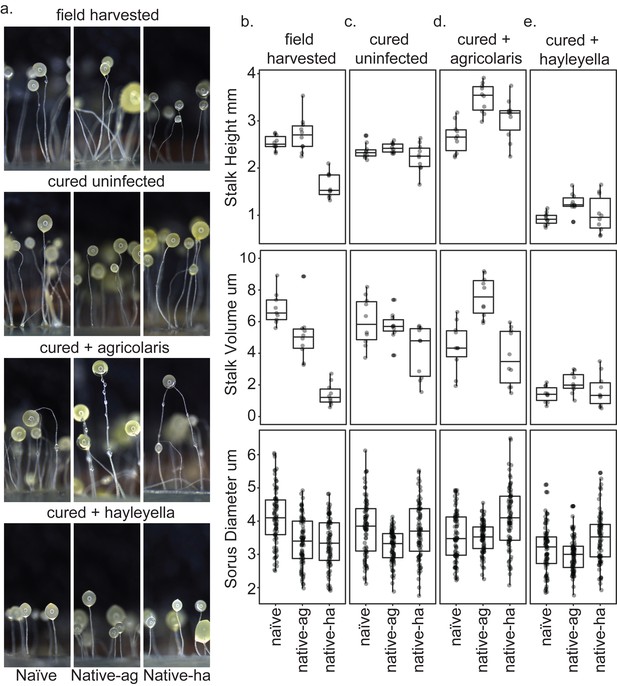
Fruiting body morphology is differentially altered by Burkholderia colonization.
Macro photographs of fruiting bodies (a) show slightly different morphologies according to Burkholderia infection status. Sori measurements demonstrate that field collected native-hayleyella hosts produce shorter stalks and less voluminous sori (b). Cured hosts produce similar fruiting body measurements across host background (c). Cured hosts subsequently infected with B. agricolaris produce slightly taller stalks, which is most noticeable in cured and re-infected native-agricolaris hosts (d). Cured hosts subsequently infected with B. hayleyella all produce significantly shorter stalks with overall smaller fruiting body dimensions (e).
Tables
Dictyostelium discoideum clones used for this study.
Clones are divided into specific sets each with naive, native-ag, and native-ha field-collected counterparts. They were collected from Virginia, North Carolina, and Texas as indicated.
| Set | Clone | Status | Burkholderia | Location collected | Date collected | GPS coordinates |
|---|---|---|---|---|---|---|
| 1 | QS9 | Naïve | None | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| QS70 | Native | B. agricolaris | Texas- Houston Arboretum | Jul. 15 2004 | N 29° 46’, W 95° 27’ | |
| QS11 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ | |
| 2 | QS18 | Naïve | None | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| QS159 | Native | B. agricolaris | Virginia-Mt Lake Biological Station | May. 2008 | N 37° 21’, W 80° 31’ | |
| QS23 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Sep. 25 2000 | N 37° 21’, W 80° 31’ | |
| 3 | QS17 | Naïve | None | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| QS161 | Native | B. agricolaris | Virginia-Mt Lake Biological Station | May. 2008 | N 37° 21’, W 80° 31’ | |
| QS22 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Sep. 25 2000 | N 37° 21’, W 80° 31’ | |
| 4 | QS6 | Naïve | None | Virginia-Mt Lake Biological Station | Sep. 25 2000 | N 37° 21’, W 80° 31’ |
| NC21 | Native | B. agricolaris | North Carolina-Little Butts Gap | Oct. 1988 | N 35° 46’, W 82° 20’ | |
| QS21 | Native | B. hayleyella | Virginia-Mt Lake Biological Station | Oct. 15 2000 | N 37° 21’, W 80° 31’ |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strainbackground(Dictyostelium discoideum) | QS6 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D.discoideum) | QS9 | Douglas et al., 2011, Brock et al., 2011 | Virginia-Mt Lake Biological Station | |
| Strain, strainbackground (D. discoideum) | QS17 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake Biological Station | |
| Strain, strainbackground (D. discoideum) | QS18 | Douglas et al., 2011, Brock et al., 2011 | Virginia-Mt Lake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS11 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS21 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS22 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS23 | Douglas et al., 2011, Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS70 | Douglas et al., 2011 | Texas- HoustonArboretum | |
| Strain, strainbackground (D. discoideum) | QS159 | Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | QS161 | Brock et al., 2011 | Virginia-MtLake BiologicalStation | |
| Strain, strainbackground (D. discoideum) | NC21 | Francis and Eisenberg, 1993 | NC-Little ButtsGap | |
| Strain, strainbackground (Burkholderia hayleyella) | BhQS11 | Haselkorn et al., 2018 | isolated fromQS11 | |
| Strain, strainbackground (B. hayleyella) | BhQS21 | Haselkorn et al., 2018 | isolated fromQS21 | |
| Strain, strainbackground (B. hayleyella) | BhQS22 | Haselkorn et al., 2018 | isolated from QS22 | |
| Strain, strainbackground (B. hayleyella) | BhQS23 | Haselkorn et al., 2018 | isolated fromQS23 | |
| Strain, strainbackground (Burkholderia agricolaris) | BaQS70 | Haselkorn et al., 2018 | isolated fromQS70 | |
| Strain, strainbackground (B. agricolaris) | BaQS159 | Haselkorn et al., 2018 | isolated fromQS159 | |
| Strain, strainbackground (B. agricolaris) | BaQS161 | Haselkorn et al., 2018 | isolated fromQS161 | |
| Strain, strain background (B. agricolaris) | BaNC21 | Haselkorn et al., 2018 | isolated fromNC21 | |
| Strain, strain background (B. agricolaris) | BaQS70-RFP.1 | DiSalvo et al., 2015 | modified fromBaQS70 | |
| Strain, strain background (B. hayleyella) | BhQS11-RFP.2 | This paper | modified fromBaQS11 | |
| Strain, strainbackground (Klebsiella pneumoniae) | KpQS | Dictybase (http://dictybase.org/) | ||
| Strain, strainbackground (K. pneumoniae) | KpQS-GFP.1 | This paper | ||
| Recombinant DNA reagent | pmini-Tn7-KS-GFP | Teal et al., 2006 | ||
| Recombinant DNA reagent | pmini-Tn7-gat-P1-RFP | Su et al., 2014 |
Additional files
-
Supplementary file 1
Statistical results of three fitness measures assayed for field-collected amoeba clones and after curing with antibiotics.
The three fitness measures were percent of spores that were viable, the total number of spores produced by a clone, and total viable spores. Total viable spores is the product of the other two measures. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Each of the fitness measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. Details about the statistical tests used can be found in the main text.
- https://doi.org/10.7554/eLife.42660.014
-
Supplementary file 2
Statistical results of three fitness measures assayed for antibiotic-cured amoeba clones after experimental addition of Burkholderia.
he three fitness measures were again percent of spores that were viable, the total number of spores produced by a clone, and total viable spores. Total viable spores is the product of the other two measures. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Each of the fitness measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. Details about the statistical tests used can be found in the main text.
- https://doi.org/10.7554/eLife.42660.015
-
Supplementary file 3
Statistical results for stalk morphology.
Each of the stalk measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Details about the statistical tests used can be found in the main text. One clone of each native type was tested: QS9 naïve; QS70 ag-infected; QS11 ha-infected.
- https://doi.org/10.7554/eLife.42660.016
-
Supplementary file 4
Statistical results for sorus morphology.
Each of the spore measures was analyzed with a set of Generalized Linear Mixed Models (GLMMs). This table gives the p-values for each question asked about main or interaction effects and the post hoc pairwise comparisons made, as relevant. For each pairwise contrast, the essential difference in treatments is in boldface, and a treatment that is significantly higher than the other is marked with an asterisk and printed in red. Details about the statistical tests used can be found in the main text. One clone of each native type was tested: QS9 naïve; QS70 ag-infected; QS11 ha-infected.
- https://doi.org/10.7554/eLife.42660.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42660.018





