Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping
Figures
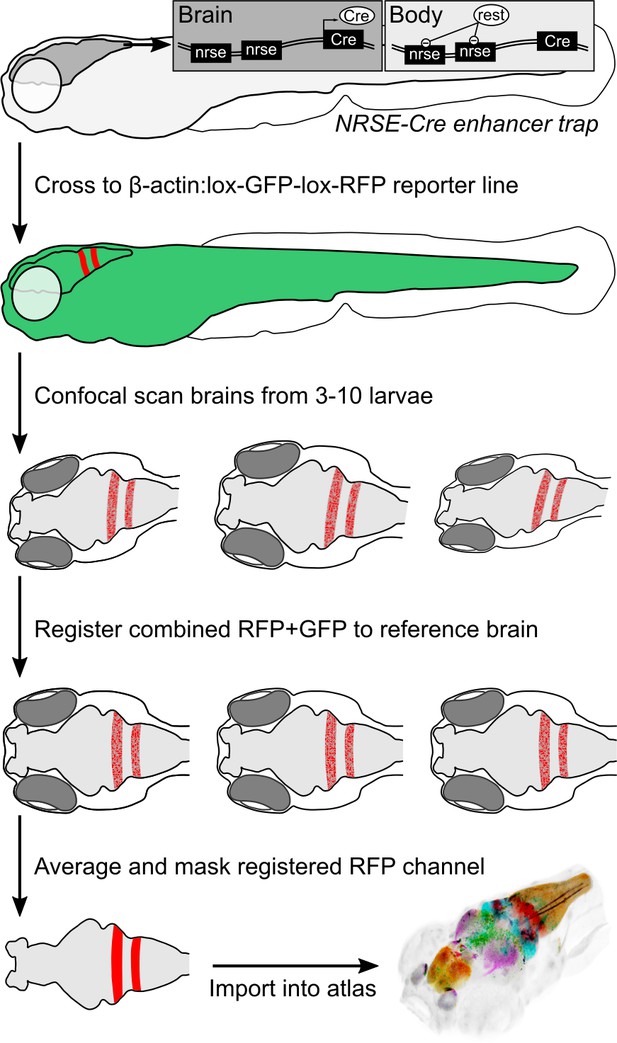
Procedure for imaging and co-registering new Cre lines.
Inset schematics: neuronal-restrictive silencing element (NRSE) sites in the enhancer trap construct are targets for the REST protein, which suppresses Cre expression outside the brain.
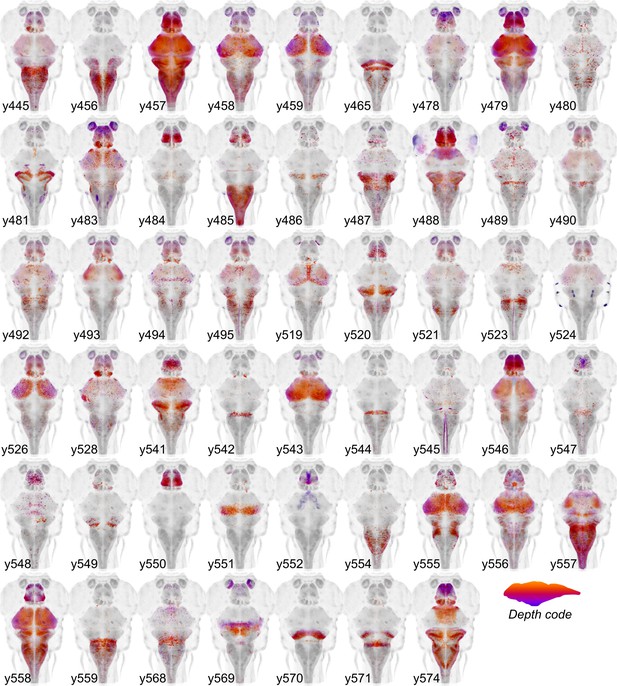
Cre enhancer trap lines.
Horizontal maximum projection of 52 new Cre enhancer trap lines, with color indicating depth along the dorsal-ventral dimension (huC counter-label, grey).
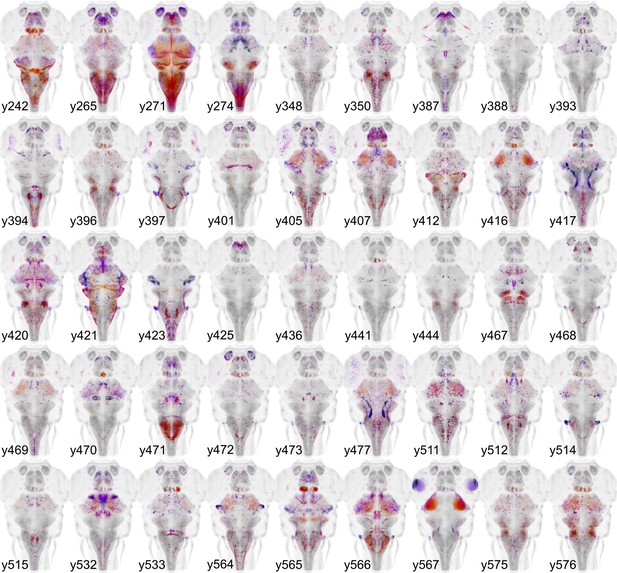
Gal4 enhancer trap lines.
Horizontal maximum projection of 45 new Gal4 enhancer trap lines (depth coded; huC counter-label, grey).
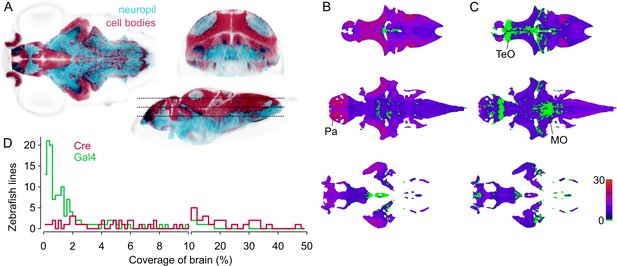
Spatial coverage of Cre and Gal4 enhancer trap lines.
(A) Horizontal (left), coronal (top right), sagittal (bottom right) sections showing huC:nls-mCar (cell bodies, red) and huC:Gal4,UAS:syp-RFP (neuropil, cyan) from ZBB (huC counter-label, gray), illustrating the separation of cell bodies and neuropil in six dpf brains. (B–C) Horizontal sections (at the levels indicated in A) of heat-maps showing the number of et-Cre (B) and et-Gal4 (C) lines that label each voxel within the cellular area of the brain (scale bar, right). Voxels that lack coverage indicated in green. Coverage for Cre lines is highest in the pallium (Pa). Gal4 lines conspicuously lack coverage in the anterior optic tectum (TeO) and in a medial zone of the medulla oblongata (MO). (D) Histogram of the cellular-region coverage for et-Cre (red) and et-Gal4 (green) lines.
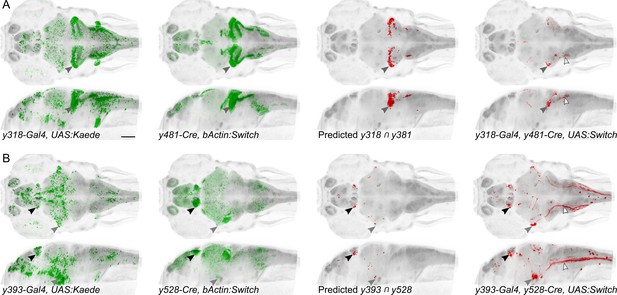
Spatially restricted reporter expression in Gal4/Cre intersectional domains.
Maximum projections for et-Gal4 and et-Cre lines predicted to show co-expression in small domains of neurons, and of the resulting pattern of expression in a triple transgenic et-Gal4, et-Cre, UAS:KillSwitch larva. Scale bar 100 µm. (A) Horizontal (top) and sagittal (bottom) maximum projections for (left to right): y318-Gal4 expression, y481-Cre expression, predicted reporter distribution in cells that co-express y318 and y481, actual RFP expression in an individual y318, y481, UAS:KillSwitch larva. Closed arrowhead indicates the cerebellar eminentia granularis (Eg), which is labeled in both lines and the intersect. Open arrowhead indicates descending fiber tract from Eg neurons. (B) As for (A) with y393-Gal4 and y528-Cre lines. Closed arrowheads indicate the lateral habenula (black) and trigeminal ganglion (grey). Open arrowhead indicates central projections from the trigeminal ganglion.
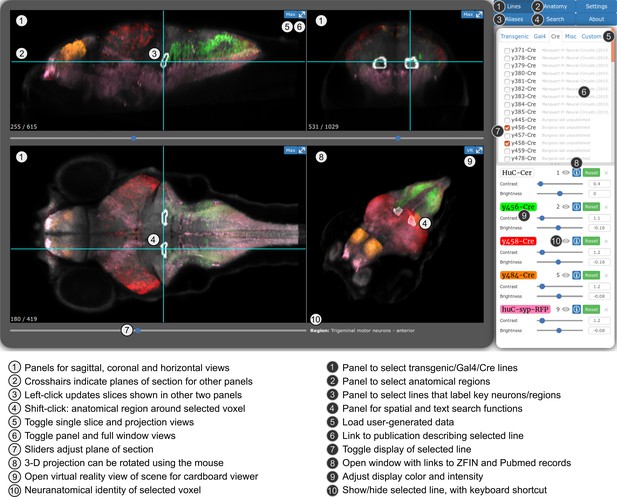
Web-browser based edition of the Zebrafish Brain Browser with key functions annotated.
Panel size can be adjusted to best fit screen dimensions with the Settings menu.
Tables
Summary of transgenic lines in ZBB2.
Total numbers of enhancer trap lines, and transgenic lines (made using promoter fragments from genes, or through BAC recombination), broken down by type: Gal4, Cre or fluorescent protein (FP). Right columns total the number of lines where genomic information driving the expression pattern is available. This information inherently exists for transgenic lines, and was derived through integration site mapping for enhancer trap lines.
| All lines (n = 264) | Mapped (n = 171) | ||||||
|---|---|---|---|---|---|---|---|
| Gal4 | Cre | FP | Gal4 | Cre | FP | ||
| Enhancer trap | 138 | 65 | 5 | 96 | 15 | 4 | |
| Transgenic | 20 | 0 | 36 | 20 | 0 | 36 | |
| Total | 158 | 65 | 41 | 116 | 15 | 40 | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. rerio) | y234 | PMID:23100441 | ZFIN:ZDB-ALT-121114–10 | ZFIN symbol:y234Et; Et(SCP1:Gal4ff)y234 |
| Genetic reagent (D. rerio) | y236 | PMID:23293587 | ZFIN:ZDB-ALT-130214–2 | ZFIN symbol:y236Et; Et(REx2-cfos:kGal4ff)y236 |
| Genetic reagent (D. rerio) | y237 | PMID:23293587 | ZFIN:ZDB-ALT-130214–3 | ZFIN symbol:y237Et; Et(REx2-cfos:kGal4ff)y237 |
| Genetic reagent (D. rerio) | y241 | PMID:24203884 | ZFIN:ZDB-ALT-131007–1 | ZFIN symbol:y241Et; Et(REx2-SCP1:kGal4ff)y241 |
| Genetic reagent (D. rerio) | y242 | This paper | ZFIN:ZDB-ALT-130214–5 | ZFIN symbol:y242Et; Et(REx2-SCP1:Gal4)y242 |
| Genetic reagent (D. rerio) | y244 | PMID:23293587 | ZFIN:ZDB-ALT-130214–7 | ZFIN symbol:y244Et; Et(REx2-SCP1:kGal4ff)y244 |
| Genetic reagent (D. rerio) | y252 | PMID:25224259 | ZFIN:ZDB-ALT-151117–1 | ZFIN symbol:y252Et; Et(REx2-SCP1:kGal4ff)y252 |
| Genetic reagent (D. rerio) | y264 | PMID:24848468 | ZFIN:ZDB-ALT-141111–2 | ZFIN symbol:y264Et; Et(SCP1:Gal4ff)y264 |
| Genetic reagent (D. rerio) | y265 | This paper | ZFIN:ZDB-ALT-180717–10 | ZFIN symbol:y265Et; Et(SCP1:Gal4ff)y265 |
| Genetic reagent (D. rerio) | y269 | PMID:24848468 | ZFIN:ZDB-ALT-141111–3 | ZFIN symbol:y269Et; Et(REx2-cfos:kGal4ff)y269 |
| Genetic reagent (D. rerio) | y270 | PMID:24848468 | ZFIN:ZDB-ALT-141111–4 | ZFIN symbol:y270Et; Et(REx2-cfos:kGal4ff)y270 |
| Genetic reagent (D. rerio) | y271 | PMID:25628360 | ZFIN:ZDB-ALT-150721–4 | ZFIN symbol:y271Et; Et(SCP1:kGal4ff)y271 |
| Genetic reagent (D. rerio) | y274 | This paper | ZFIN:ZDB-ALT-180717–11 | ZFIN symbol:y274Et; Et(SCP1:Gal4ff)y274 |
| Genetic reagent (D. rerio) | y348 | This paper | ZFIN:ZDB-ALT-180717–12 | ZFIN symbol:y348Et; Et(REx2-SCP1:kGal4ff)y348 |
| Genetic reagent (D. rerio) | y387 | This paper | ZFIN:ZDB-ALT-180717–14 | ZFIN symbol:y387Et; Et(tph2:Gal4ff)y387 |
| Genetic reagent (D. rerio) | y394 | This paper | ZFIN:ZDB-ALT-180717–16 | ZFIN symbol:y394Et; Et(cfos:Gal4ff)y394 |
| Genetic reagent (D. rerio) | y396 | PMID:26635538 | ZFIN:ZDB-ALT-170320–13 | ZFIN symbol:y396Et; Et(SCP1:Gal4ff)y396 |
| Genetic reagent (D. rerio) | y412 | This paper | ZFIN:ZDB-ALT-180717–20 | ZFIN symbol:y412Et; Et(cfos:Gal4ff)y412 |
| Genetic reagent (D. rerio) | y416 | This paper | ZFIN:ZDB-ALT-180717–21 | ZFIN symbol:y416Et; Et(cfos:Gal4ff)y416 |
| Genetic reagent (D. rerio) | y420 | PMID:26635538 | ZFIN:ZDB-ALT-180717–23 | ZFIN symbol:y420Et; Et(REx2-cfos:Gal4ff)y420 |
| Genetic reagent (D. rerio) | y421 | This paper | ZFIN:ZDB-ALT-180717–24 | ZFIN symbol:y421Et; Et(REx2-cfos:Gal4)y421 |
| Genetic reagent (D. rerio) | y433 | This paper | ZFIN:ZDB-ALT-151125–10 | ZFIN symbol:y433Et; Et(cfos:kGal4ff)y433 |
| Genetic reagent (D. rerio) | y436 | This paper | ZFIN:ZDB-ALT-151125–13 | ZFIN symbol:y436Et; Et(cfos:Gal4ff)y436 |
| Genetic reagent (D. rerio) | y441 | This paper | ZFIN:ZDB-ALT-180717–27 | ZFIN symbol:y441Et; Et(cfos:Gal4ff)y441 |
| Genetic reagent (D. rerio) | y444 | This paper | ZFIN:ZDB-ALT-180717–28 | ZFIN symbol:y444Et; Et(SCP1:Gal4ff)y444 |
| Genetic reagent (D. rerio) | y445 | This paper | ZFIN:ZDB-ALT-180717–29 | ZFIN symbol:y445Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y445 |
| Genetic reagent (D. rerio) | y456 | This paper | ZFIN:ZDB-ALT-180717–30 | ZFIN symbol:y456Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y456 |
| Genetic reagent (D. rerio) | y457 | This paper | ZFIN:ZDB-ALT-180717–31 | ZFIN symbol:y457Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y457 |
| Genetic reagent (D. rerio) | y458 | This paper | ZFIN:ZDB-ALT-180717–32 | ZFIN symbol:y458Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y458 |
| Genetic reagent (D. rerio) | y459 | This paper | ZFIN:ZDB-ALT-180717–33 | ZFIN symbol:y459Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y459 |
| Genetic reagent (D. rerio) | y465 | This paper | ZFIN:ZDB-ALT-180717–34 | ZFIN symbol:y465Et; Et(REx2-SCP1:BGi-Cre-2a-Cer)y465 |
| Genetic reagent (D. rerio) | y467 | This paper | ZFIN:ZDB-ALT-180717–35 | ZFIN symbol:y467Et; Et(tph2:Gal4ff)y467 |
| Genetic reagent (D. rerio) | y468 | This paper | ZFIN:ZDB-ALT-180717–36 | ZFIN symbol:y468Et; Et(cfos:Gal4ff)y468 |
| Genetic reagent (D. rerio) | y469 | This paper | ZFIN:ZDB-ALT-180717–37 | ZFIN symbol:y469Et; Et(cfos:Gal4ff)y469 |
| Genetic reagent (D. rerio) | y470 | This paper | ZFIN:ZDB-ALT-180717–38 | ZFIN symbol:y470Et; Et(REx2-SCP1:Gal4ff)y470 |
| Genetic reagent (D. rerio) | y471 | This paper | ZFIN:ZDB-ALT-180717–39 | ZFIN symbol:y471Et; Et(REx2-SCP1:Gal4ff)y471 |
| Genetic reagent (D. rerio) | y472 | This paper | ZFIN:ZDB-ALT-180717–40 | ZFIN symbol:y472Et; Et(REx2-SCP1:Gal4ff)y472 |
| Genetic reagent (D. rerio) | y473 | This paper | ZFIN:ZDB-ALT-180717–41 | ZFIN symbol:y473Et; Et(cfos:Gal4ff)y473 |
| Genetic reagent (D. rerio) | y477 | This paper | ZFIN:ZDB-ALT-180717–43 | ZFIN symbol:y477Et; Et(cfos:Gal4ff)y477 |
| Genetic reagent (D. rerio) | y478 | This paper | ZFIN:ZDB-ALT-180717–44 | ZFIN symbol:y478Et; Et(attP-REx2-SCP1:BGi-Cre-2a-Cer-attP)y478 |
| Genetic reagent (D. rerio) | y479 | This paper | ZFIN:ZDB-ALT-180717–45 | ZFIN symbol:y479Et; Et(attP-REx2-SCP1:BGi-Cre-2a-Cer-attP)y479 |
| Genetic reagent (D. rerio) | y480 | This paper | ZFIN:ZDB-ALT-180717–46 | ZFIN symbol:y480Et; Et(attP-REx2-SCP1:BGi-Cre-2a-Cer-attP)y480 |
| Genetic reagent (D. rerio) | y481 | This paper | ZFIN:ZDB-ALT-180717–47 | ZFIN symbol:y481Et; Et(attP-REx2-SCP1:BGi-Cre-2a-Cer-attP)y481 |
| Genetic reagent (D. rerio) | y483 | This paper | ZFIN:ZDB-ALT-180717–48 | ZFIN symbol:y483Et; Et(attP-REx2-SCP1:BGi-Cre-2a-Cer-attP)y483 |
| Genetic reagent (D. rerio) | y484 | This paper | ZFIN:ZDB-ALT-180717–49 | ZFIN symbol:y484Et; Et(REx2-SCP1:BGi-Cre-2a-Cer)y484 |
| Genetic reagent (D. rerio) | y485 | This paper | ZFIN:ZDB-ALT-180717–50 | ZFIN symbol:y485Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y485 |
| Genetic reagent (D. rerio) | y486 | This paper | ZFIN:ZDB-ALT-180717–51 | ZFIN symbol:y486Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y486 |
| Genetic reagent (D. rerio) | y487 | This paper | ZFIN:ZDB-ALT-180717–52 | ZFIN symbol:y487Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y487 |
| Genetic reagent (D. rerio) | y488 | This paper | ZFIN:ZDB-ALT-180717–53 | ZFIN symbol:y488Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y488 |
| Genetic reagent (D. rerio) | y489 | This paper | ZFIN:ZDB-ALT-180717–54 | ZFIN symbol:y489Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y489 |
| Genetic reagent (D. rerio) | y490 | This paper | ZFIN:ZDB-ALT-180717–55 | ZFIN symbol:y490Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y490 |
| Genetic reagent (D. rerio) | y492 | This paper | ZFIN:ZDB- ALT-180717–56 | ZFIN symbol:y492Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y492 |
| Genetic reagent (D. rerio) | y493 | This paper | ZFIN:ZDB-ALT-180717–57 | ZFIN symbol:y493Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y493 |
| Genetic reagent (D. rerio) | y494 | This paper | ZFIN:ZDB-ALT-180717–58 | ZFIN symbol:y494Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y494 |
| Genetic reagent (D. rerio) | y495 | This paper | ZFIN:ZDB-ALT-180717–59 | ZFIN symbol:y495Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y495 |
| Genetic reagent (D. rerio) | y511 | This paper | ZFIN:ZDB-ALT-180717–60 | ZFIN symbol:y511Et; Et(cfos:Gal4ff)y511 |
| Genetic reagent (D. rerio) | y512 | This paper | ZFIN:ZDB-ALT-180717–61 | ZFIN symbol:y512Et; Et(SCP1:Gal4)y512 |
| Genetic reagent (D. rerio) | y514 | This paper | ZFIN:ZDB-ALT-180717–63 | ZFIN symbol:y514Et; Et(REx2-cfos:kGal4ff)y514 |
| Genetic reagent (D. rerio) | y515 | This paper | ZFIN:ZDB-ALT-180717–64 | ZFIN symbol:y515Et; Et(REx2-cfos:kGal4ff)y515 |
| Genetic reagent (D. rerio) | y519 | This paper | ZFIN:ZDB-ALT-180717–65 | ZFIN symbol:y519Et; Et(REx2-SCP1:BGi-Cre-2a-Cer)y519 |
| Genetic reagent (D. rerio) | y520 | This paper | ZFIN:ZDB-ALT-180717–66 | ZFIN symbol:y520Et; Et(REx2-SCP1:BGi-Cre-2a-Cer)y520 |
| Genetic reagent (D. rerio) | y521 | This paper | ZFIN:ZDB-ALT-180717–67 | ZFIN symbol:y521Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y521 |
| Genetic reagent (D. rerio) | y523 | This paper | ZFIN:ZDB-ALT-180717–68 | ZFIN symbol:y523Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y523 |
| Genetic reagent (D. rerio) | y524 | This paper | ZFIN:ZDB-ALT-180717–69 | ZFIN symbol:y524Et; Et(REx2-SCP1:BGi-Cre-2a-Cer)y524 |
| Genetic reagent (D. rerio) | y526 | This paper | ZFIN:ZDB-ALT-180717–70 | ZFIN symbol:y526Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y526 |
| Genetic reagent (D. rerio) | y528 | This paper | ZFIN:ZDB-ALT-180717–71 | ZFIN symbol:y528Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y528 |
| Genetic reagent (D. rerio) | y532 | This paper | ZFIN:ZDB-ALT-180717–72 | ZFIN symbol:y532Et; Et(cfos:Gal4ff)y532 |
| Genetic reagent (D. rerio) | y533 | This paper | ZFIN:ZDB-ALT-180717–73 | ZFIN symbol:y533Et; Et(SCP1:Gal4ff)y533 |
| Genetic reagent (D. rerio) | y541 | This paper | ZFIN:ZDB-ALT-180717–74 | ZFIN symbol:y541Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y541 |
| Genetic reagent (D. rerio) | y542 | This paper | ZFIN:ZDB-ALT-180717–75 | ZFIN symbol:y542Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y542 |
| Genetic reagent (D. rerio) | y543 | This paper | ZFIN:ZDB-ALT-180717–76 | ZFIN symbol:y543Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y543 |
| Genetic reagent (D. rerio) | y544 | This paper | ZFIN:ZDB-ALT-180717–77 | ZFIN symbol:y544Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y544 |
| Genetic reagent (D. rerio) | y545 | This paper | ZFIN:ZDB-ALT-180717–78 | ZFIN symbol:y545Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y545 |
| Genetic reagent (D. rerio) | y546 | This paper | ZFIN:ZDB-ALT-180717–79 | ZFIN symbol:y546Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y546 |
| Genetic reagent (D. rerio) | y547 | This paper | ZFIN:ZDB-ALT-180717–80 | ZFIN symbol:y547Et; Et(REx2-SCP1:BGi-Cre)y547 |
| Genetic reagent (D. rerio) | y548 | This paper | ZFIN:ZDB-ALT-180717–81 | ZFIN symbol:y548Et; Et(REx2-SCP1:BGi-Cre)y548 |
| Genetic reagent (D. rerio) | y549 | This paper | ZFIN:ZDB-ALT-180717–82 | ZFIN symbol:y549Et; Et(REx2-SCP1:BGi-Cre)y549 |
| Genetic reagent (D. rerio) | y550 | This paper | ZFIN:ZDB-ALT-180717–83 | ZFIN symbol:y550Et; Et(REx2-SCP1:BGi-Cre)y550 |
| Genetic reagent (D. rerio) | y551 | This paper | ZFIN:ZDB-ALT-180717–84 | ZFIN symbol:y551Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y551 |
| Genetic reagent (D. rerio) | y552 | This paper | ZFIN:ZDB-ALT-180717–85 | ZFIN symbol:y552Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y552 |
| Genetic reagent (D. rerio) | y554 | This paper | ZFIN:ZDB-ALT-180717–87 | ZFIN symbol:y554Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y554 |
| Genetic reagent (D. rerio) | y555 | This paper | ZFIN:ZDB-ALT-180717–88 | ZFIN symbol:y555Et; Et(REx2-SCP1:BGi-Cre)y555 |
| Genetic reagent (D. rerio) | y556 | This paper | ZFIN:ZDB-ALT-180717–89 | ZFIN symbol:y556Et; Et(REx2-SCP1:BGi-Cre)y556 |
| Genetic reagent (D. rerio) | y557 | This paper | ZFIN:ZDB-ALT-180717–90 | ZFIN symbol:y557Et; Et(REx2-SCP1:BGi-Cre)y557 |
| Genetic reagent (D. rerio) | y558 | This paper | ZFIN:ZDB-ALT-180717–91 | ZFIN symbol:y558Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y558 |
| Genetic reagent (D. rerio) | y559 | This paper | ZFIN:ZDB-ALT-180717–92 | ZFIN symbol:y559Et; Et(REx2-SCP1:BGi-Cre)y559 |
| Genetic reagent (D. rerio) | y564 | This paper | ZFIN:ZDB-ALT-180717–93 | ZFIN symbol:y564Et; Et(SCP1:Gal4ff)y564 |
| Genetic reagent (D. rerio) | y565 | This paper | ZFIN:ZDB-ALT-180717–94 | ZFIN symbol:y565Et; Et(REx2-SCP1:Gal4ff)y565 |
| Genetic reagent (D. rerio) | y566 | This paper | ZFIN:ZDB-ALT-180717–95 | ZFIN symbol:y566Et; Et(REx2-cfos:Gal4ff)y566 |
| Genetic reagent (D. rerio) | y567 | This paper | ZFIN:ZDB-ALT-180717–96 | ZFIN symbol:y567Et; Et(REx2-cfos:Gal4ff)y567 |
| Genetic reagent (D. rerio) | y568 | This paper | ZFIN:ZDB-ALT-180717–97 | ZFIN symbol:y568Et; Et(REx2-SCP1:BGi-Cre-2a-Cer.zf3)y568 |
| Genetic reagent (D. rerio) | y569 | This paper | ZFIN:ZDB-ALT-180717–98 | ZFIN symbol:y569Et; Et(REx2-SCP1:BGi-Cre)y569 |
| Genetic reagent (D. rerio) | y570 | This paper | ZFIN:ZDB-ALT-180717–99 | ZFIN symbol:y570Et; Et(REx2-SCP1:BGi-Cre)y570 |
| Genetic reagent (D. rerio) | y571 | This paper | ZFIN:ZDB-ALT-180717–100 | ZFIN symbol:y571Et; Et(R2R6-hoxa2-CNE-SCP1:BGi-Cre-2a-Cer)y571 |
| Genetic reagent (D. rerio) | y574 | This paper | ZFIN:ZDB-ALT-180717–101 | ZFIN symbol:y574Et; Et(REx2-SCP1:BGi-Cre)y574 |
| Genetic reagent (D. rerio) | y575 | This paper | ZFIN:ZDB-ALT-180717–102 | ZFIN symbol:y575Et; Et(cfos:Gal4)y575 |
| Genetic reagent (D. rerio) | y576 | This paper | ZFIN:ZDB-ALT-180717–103 | ZFIN symbol:y576Et; Et(cfos:Gal4)y576 |
| Genetic reagent (D. rerio) | βactin:Switch; y272 | PMID:25628360 | ZFIN:ZDB-ALT-150721–8 | |
| Genetic reagent (D. rerio) | vglut2a:DsRed; nns14 | PMID:22302816 | ZFIN:ZDB-ALT-110413–5 | |
| Genetic reagent (D. rerio) | UAS:KillSwitch; y518 | PMID:30078569 | ZFIN:ZDB-ALT-181218–5 | |
| Genetic reagent (D. rerio) | UAS:Kaede; s1999t | PMID:17335798 | ZFIN:ZDB-ALT-070314–1 | |
| Genetic reagent (D. rerio) | elavl3:h2b-GCaMP6; jf5 | PMID:25068735 | ZFIN:ZDB-ALT-141023–2 | |
| Recombinant DNA reagent | REx2-SCP1:BGi-Cre-2a-Cer (plasmid) | PMID:26635538 | ZFIN:ZDB-ETCONSTRCT-151102–1 | |
| Recombinant DNA reagent | REx2-SCP1:BGi-Cre | This paper | ZFIN:ZDB-ETCONSTRCT-180514–2 | Progenitors:REx2-SCP1:BGi-Cre-2a-Cer (plasmid) |
| Recombinant DNA reagent | REx2-SCP1:BGi-Cre-2a-Cer.zf3 (plasmid) | This paper | ZFIN:ZDB-ETCONSTRCT-180518–1 | Progenitors:REx2-SCP1:BGi-Cre-2a-Cer (plasmid) |
| Recombinant DNA reagent | attP-REx2-SCP1:BGi-Cre-2a-Cer-attP | This paper | ZFIN:ZDB-ETCONSTRCT-151102–2 | Progenitors:REx2-SCP1:BGi-Cre-2a-Cer (plasmid) |
| Recombinant DNA reagent | R2R6-hoxa2-CNE-SCP1:BGi-Cre-2a-Cer | This paper | ZFIN:ZDB-ETCONSTRCT-180514–1 | Progenitors:REx2-SCP1:BGi-Cre-2a-Cer (plasmid) |
| Sequence-based reagent | tol2-arrm pulldown oligonucleotide | This paper | 5-CTCAAGTGAAAGTACAAGTACTTAGGGAAAATTTTACTCAATTAAAAGTAAAAGTATCTGGCTAGAATCTTACTTGAGTAAAAGTAAAAAAGTACTCCATTAAAATTGTACTTGAGTATT | |
| Sequence-based reagent | tol2-arrm pulldown oligonucleotide | This paper | 5-TGTAATTAAGTAAAAGTAAAAGTATTGATTTTTAATTGTACTCAAGTAAAGTAAAAATCCCCAAAAATAATACTTAAGTACAGTAATCAAGTAAAATTACTCAAGTACTTTACACCTCTG | |
| Software | Advanced Normalization Tools | PMID:17659998 | ||
| Software | Zebrafish Brain Browser (desktop) | PMID:26635538 | Download at https://science.nichd.nih.gov/confluence/display/burgess/Brain+Browser | |
| Software | Zebrafish Brain Browser (online) | This paper | GitHub:BurgessLab/ZebrafishBrainBrowser | zbbrowser.com; Githhub hosts javascript code using X3DOM to render image files |
| Software | ImageJ | PMID:22930834 | ||
| Other | Confocal images for ZBB2 lines | This paper | Dryad:doi:10.5061/dryad.tk467n8 | Compressed archives containing 16-bit NIFTI format scans of individual larvae |
Additional files
-
Supplementary file 1
Summary of Cre lines generated by our lab, available in ZBB2.
Integration site given in zv10 coordinates, transgene orientation indicated in brackets.
Integration sites inside genes are annotated, or nearest Refseq gene within ~50 kb indicated.
- https://doi.org/10.7554/eLife.42687.009
-
Supplementary file 2
Summary of all Gal4 lines generated by our lab, available in ZBB2.
Coordinate system for mapped lines is either zv9 or zv10 as indicated.
- https://doi.org/10.7554/eLife.42687.010
-
Supplementary file 3
Transgene expression intensity in small volumes within the brain.
First three columns provide central coordinates of 20 × 20 × 20 µm volumes in the ZBB reference volume (Horizontal plane: distance in microns from the dorsal surface of the volume; Transverse plane: distance from the anterior edge of the volume; Sagittal plane: distance from the left side of the volume). Additional columns indicate the mean expression per bin of each transgene after scaling voxels from 0 to 1.0.
- https://doi.org/10.7554/eLife.42687.011
-
Supplementary file 4
Cell-type expression levels in neuroanatomical regions.
Mean expression levels of transgenic markers in neuroanatomical regions defined by Z-Brain. Cell type markers: Glutamatergic (vglut2a:DsRed), GABAergic (gad1b:GFP), Glycinergic (glyt2:GFP), Catecholaminergic (vmat2:GFP), Serotonergic, (pet1:GFP), Cholinergic (chata:Gal4), Dopaminergic (th:Gal4), Hypocretin (hcrt:RFP), and glia (gfap:GFP).
- https://doi.org/10.7554/eLife.42687.012
-
Supplementary file 5
Cell-type expression levels in neuroanatomical regions.
Mean expression levels of transgenic markers in computationally defined neuroanatomical regions.
- https://doi.org/10.7554/eLife.42687.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42687.014





