The COMA complex interacts with Cse4 and positions Sli15/Ipl1 at the budding yeast inner kinetochore
Figures
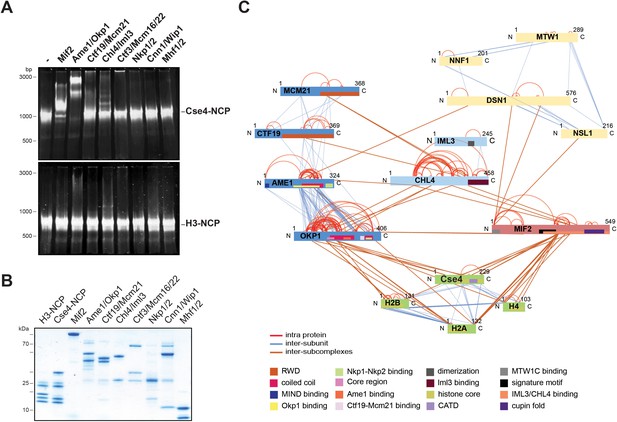
The heterodimeric Ame1/Okp1 complex directly and selectively binds the Cse4-NCP.
(A) Electrophoretic mobility shift assays (EMSAs) of the indicated CTF19cCCAN subunits and subcomplexes mixed in a 2:1 molar ratio with either Cse4- or H3-NCPs. DNA/protein complexes were separated on a 6% native polyacrylamide gel. The DNA is visualized by SYBR Gold staining. (B) Coomassie stained gel of the individual inner kinetochore components, recombinantly purified from E. coli, used in the EMSA in (A). (C) XLMS analysis of the in vitro reconstituted Cse4-NCP:Mif2:COMA:Chl4/Iml3:MTW1c complex. Proteins are represented as bars indicating annotated domains (Supplementary file 3) according to the color scheme in the legend. Subunits of a complex are represented in the same color and protein lengths and cross-link sites are scaled to the amino acid sequence.
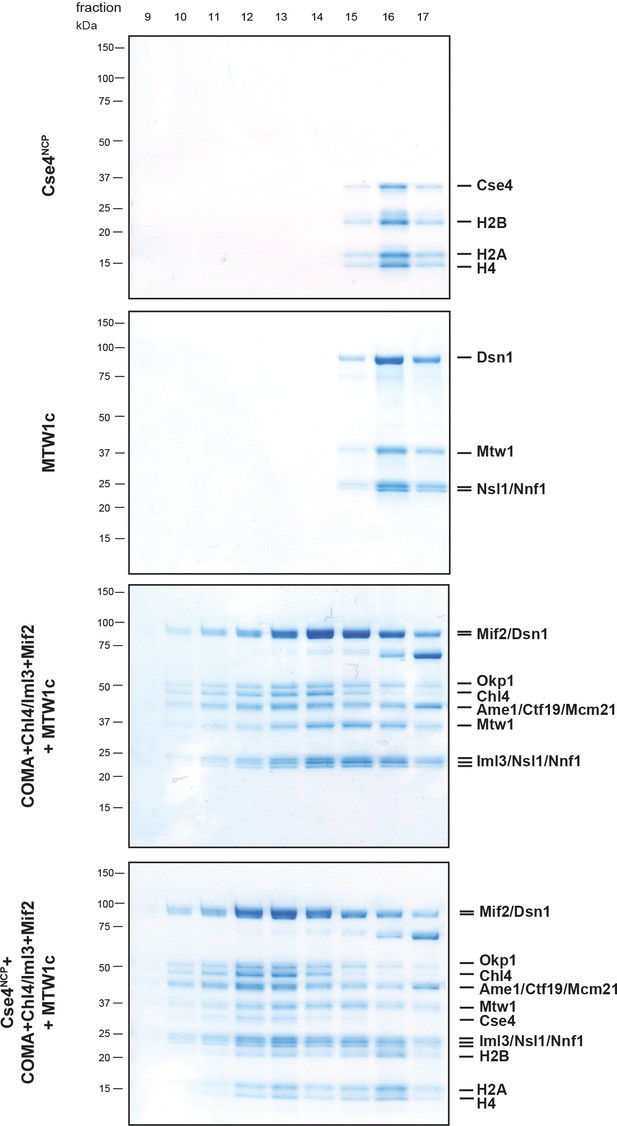
Size exclusion chromatography (SEC) of the in vitro reconstituted Ctf19/Mcm21/Ame1/Okp1 (COMA):Chl4/Iml3:Mif2:MTW1c:Cse4-NCP complex.
Proteins were mixed in an equimolar ratio, incubated on ice for 1 hr and run on a Superose 6 increase 3.2/300 column. Eluted proteins were visualized by SDS-PAGE and Coomassie staining. For the XLMS analysis shown in Figure 1C the pre-incubated complex was cross-linked prior to SEC, and elution fractions 12 and 13 corresponding to the non-cross-linked complex were pooled and digested for mass spectrometric analysis.
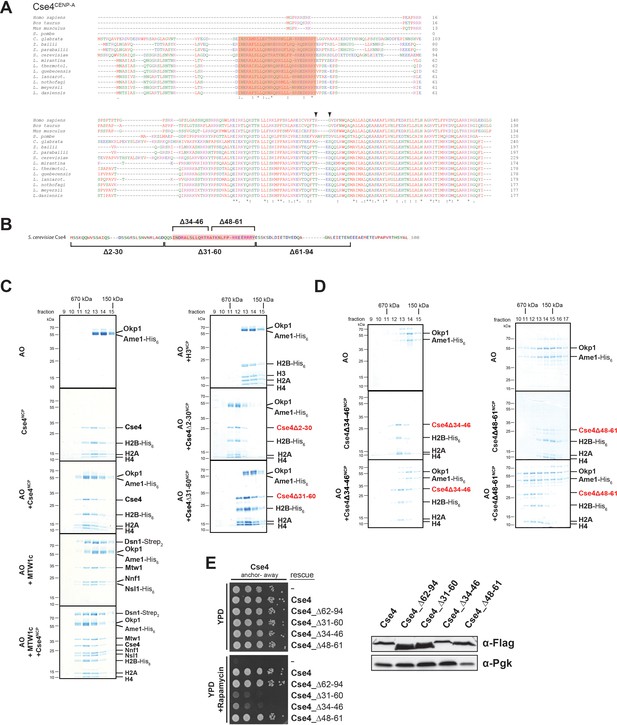
A short helical motif within the Cse4 N-terminus serves as Ame1/Okp1 docking site and is essential in vivo.
(A) Multiple sequence alignment of Cse4CENP-A proteins. Yeast protein sequences with the highest similarities to S. cerevisiae Cse4, three mammalian and the S. pombe homologous CENP-A protein sequences were included in the alignment. The amino acid (aa) patch, conserved in interrelated yeasts, is highlighted in pink (S. cerevisiae Cse4 aa 34–61). The RG motif in the mammalian sequences is indicated by arrowheads. Amino acid residues are colored and annotated according to the ClustalW color and annotation codes (S.: Schizosaccharomyces, C.: Candida, Z.: Zygosaccharomyces, L.: Lachancea). Residues that are identical among aligned protein sequences (*), conserved substitutions (:), and semiconserved substitutions (.) are indicated. (B) Scheme of the deletion mutants within the Cse4 N-terminus used in the SEC experiments in (C) and (D) and in the cell viability assays in (E). The conserved region (aa 34–61) is highlighted in pink. (C) SEC analysis of the indicated mixtures of recombinant Ame1/Okp1 (AO) and MTW1c and reconstituted H3-, Cse4-, Cse4Δ2–30- or Cse4Δ31–60-NCPs. Ame1/Okp1, MTW1c and the Cse4 proteins were mixed equimolar. Eluted proteins were visualized by SDS-PAGE and Coomassie staining. (D) SEC analysis of Ame1/Okp1 (AO) preincubated with Cse4Δ34–46- or Cse4Δ48–61-NCPs. Eluted complexes were analyzed by SDS-PAGE and Coomassie staining. (E) Left panel: Cell growth assay of Cse4 mutants in budding yeast using the anchor-away system. The Cse4 wild-type and indicated mutant proteins were ectopically expressed in a Cse4 anchor-away strain (Cse4-FRB) and cell growth was monitored by plating 1:10 serial dilutions on YPD medium at 30°C in the absence or presence of 1 µg/ml rapamycin. Right panel: Western blot analysis of the ectopically expressed Cse4 wild-type and mutant protein levels in the yeast strains shown on the left. Pgk1 levels are shown as loading control.
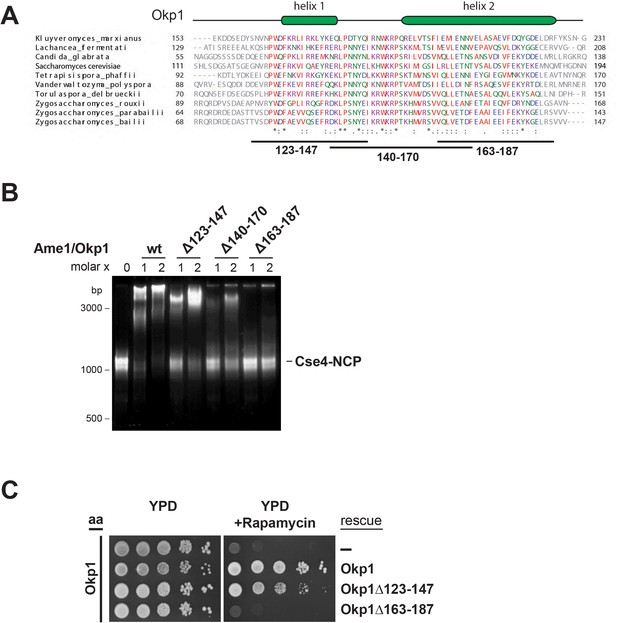
The essential core domain of Okp1 is required for the interaction with Cse4-NCPs.
(A) Multiple sequence alignment of Okp1 amino acid sequences from related yeast species. Amino acid residues of the conserved region are colored and annotated according to the ClustalW color and annotation codes. Green bars above the alignment represent alpha helical regions predicted by Jpred (Drozdetskiy et al., 2015). Lines below the alignment indicate the overlapping Okp1 deletion mutants analysed in (B) and (C). Residues that are identical among aligned protein sequences (*), conserved substitutions (:), and semiconserved substitutions (.) are indicated. (B) EMSA assessing complex formation of Cse4-NCPs with Ame1/Okp1 including wild-type (wt) Okp1 and the indicated Okp1 deletion mutants. Recombinant Ame1/Okp1 complexes were tested in a 1:1 (1) and 2:1 (2) molar ratio with Cse4-NCPs. The DNA is visualized by SYBR Gold staining. (C) Cell viability assay of Okp1 deletion mutants using the anchor away (aa) technique. Yeast growth of either the untransformed (-) Okp1 anchor-away strain (Okp1-FRB) or of strains transformed with the indicated Okp1 rescue alleles was tested in 1:10 serial dilutions on YPD medium in the absence or presence of 1 µg/ml rapamycin for 72 hr at 30°C.
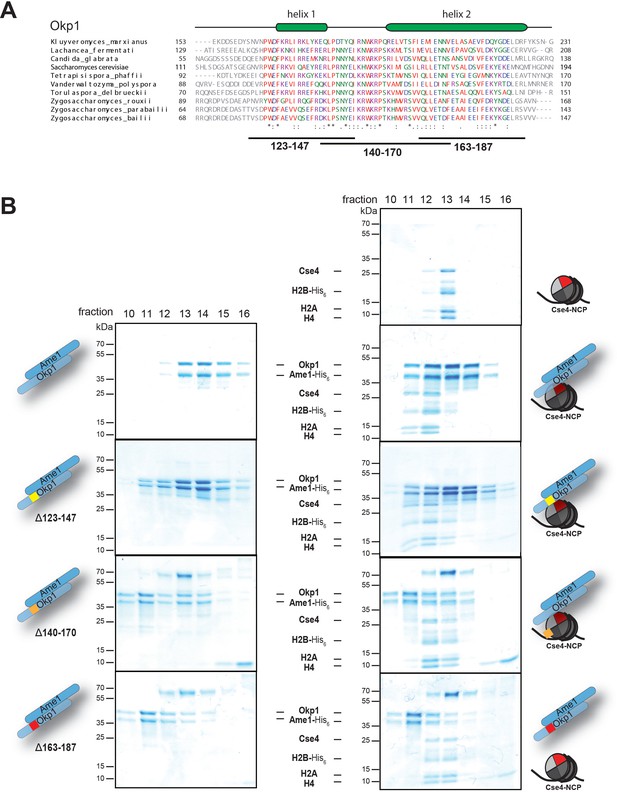
Identification of the Cse4 binding site on Okp1.
(A) Multiple sequence alignment of Okp1 amino acid sequences from related yeast species. Amino acid residues of the conserved region are colored and annotated according to the ClustalW color and annotation codes. Green bars above the alignment represent alpha helical regions predicted by Jpred (Drozdetskiy et al., 2015). Lines below the alignment indicate the overlapping Okp1 deletion mutants analysed in (B). (B) Size exclusion chromatography (SEC) analysis of equimolar mixtures of reconstituted Cse4-NCPs with recombinant wild-type Ame1/Okp1, Ame1/Okp1Δ123–147, Ame1/Okp1Δ140–170 or Ame1/Okp1Δ163–187 mutant complexes. Eluted proteins were visualized by SDS-PAGE and Coomassie staining.
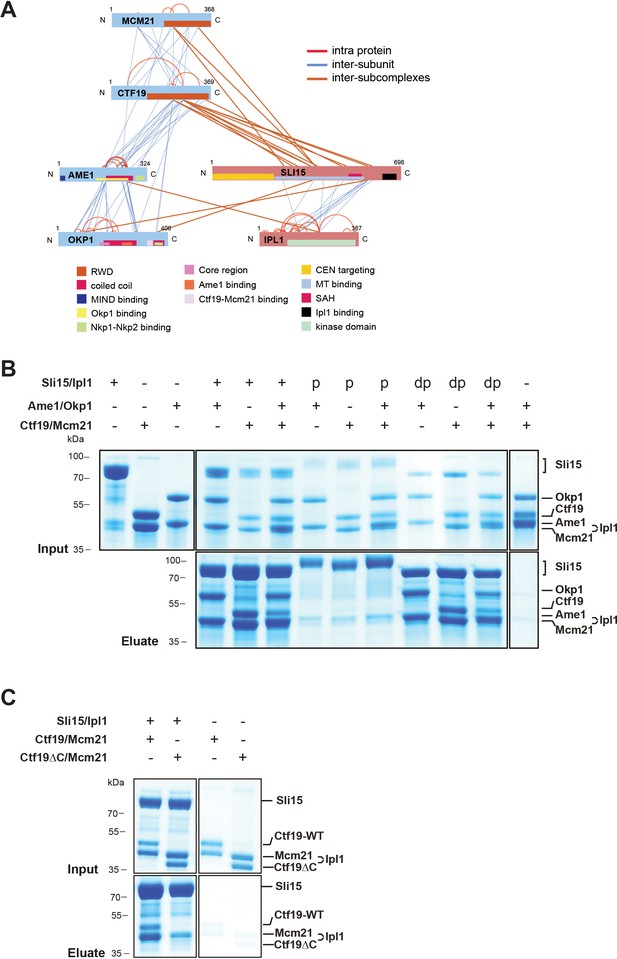
The core-CPC Sli15/Ipl1 associates with the COMA complex through the Ctf19 C-terminal RWD domain in vitro.
(A) Network representation of lysine-lysine cross-links identified on recombinant Sli15/Ipl1 in complex with COMA. Proteins are represented as bars indicating annotated domains (Supplementary file 3) according to the color scheme in the legend. Subunits of a complex are represented in the same color. Protein lengths and cross-link sites are scaled to the amino acid sequence. (B) In vitro binding assay analyzing the interaction of Sli15/Ipl1 with the COMA complex. Recombinant Sli15-2xStrep/Ipl1 was immobilized on Streptavidin beads and incubated with Ctf19/Mcm21, Ame1/Okp1 or Ame1/Okp1/Ctf19/Mcm21. Autophosphorylation (p) of Sli15/Ipl1 largely reduced bound protein levels. Dephosphorylation (dp) of Sli15/Ipl1 did not alter the bound proteins levels, which were visualized by SDS-PAGE and Coomassie staining. (C) In vitro binding assay analyzing the interaction of Sli15/Ipl1 with Ctf19/Mcm21 or Ctf19∆C/Mcm21. Ctf19∆C lacks the last 100 amino acids which form the C-terminal RWD domain. This panel is representative of three independent experiments.
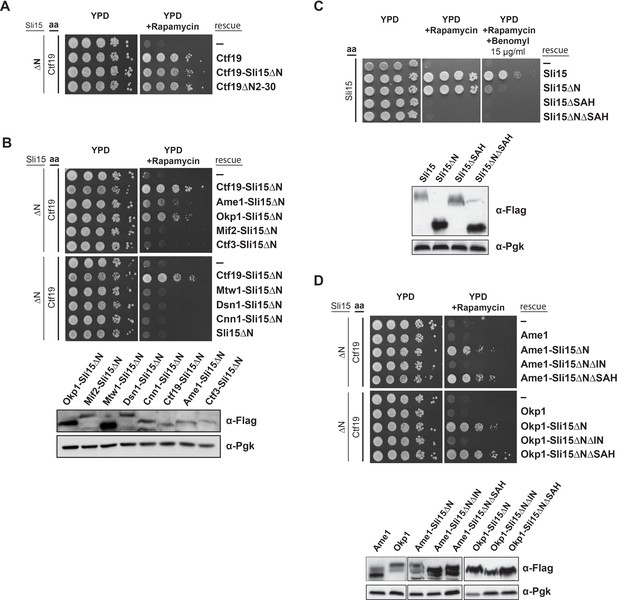
Synthetic lethality of Sli15ΔN and Ctf19 depletion is rescued by fusing Sli15ΔN to Ame1/Okp1 and is independent of Ctf19’s role in cohesin loading.
(A)-(D) Cell viability assays studying the rescue of synthetic lethality of a sli15ΔN/CTF19-FRB strain using the anchor-away system. The indicated constructs were transformed into a Ctf19 anchor-away (aa) strain (Ctf19-FRB) carrying sli15∆N (∆N) at the endogenous locus (A, B, D,) or into a Sli15 anchor-away strain (Sli15-FRB) (C). Yeast growth was tested in serial dilutions either untransformed (-) or transformed with the indicated rescue constructs on YPD medium in the absence or presence of 1 µg/ml rapamycin at 30°C. The lower panels in (B), (C) and (D) show western blot analysis of the ectopically expressed protein levels. Pgk1 levels are shown as loading control. (A) Deletion of the Ctf19 N-terminus (Ctf19∆N2-30) does not affect cell viability in a sli15∆N background. (B) Artificial tethering of Sli15∆N to Ame1 or Okp1 rescued synthetic lethality of sli15∆N cells upon Ctf19-FRB depletion from the nucleus. (C) Growth phenotypes of Sli15 wild-type, Sli15∆SAH, Sli15∆N, and Sli15∆N∆SAH tested in a Sli15-FRB anchor-away strain. (D) Rescue of cell growth by ectopic Ame1-Sli15∆N or Okp1-Sli15∆N fusion proteins is dependent on the Sli15 Ipl1-binding domain (IN-box), whereas the SAH domain is dispensable.
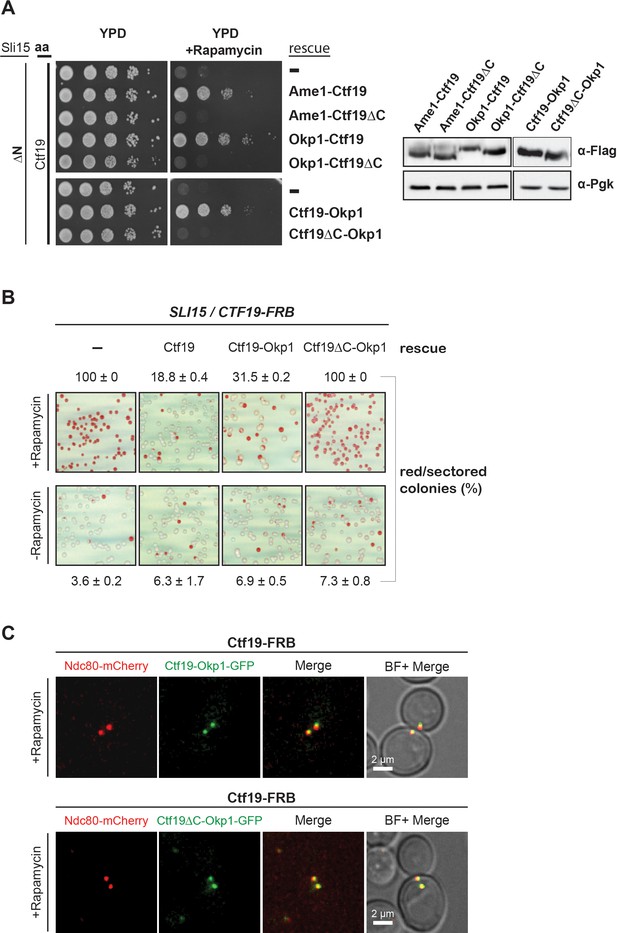
The Ctf19 C-terminus is important for chromosome segregation in the Sli15 wild-type background.
(A) Left panel: Growth assay of the sli15ΔN/CTF19-FRB strain expressing Ame1-Ctf19, Ame1-Ctf19∆C, Okp1-Ctf19, Okp1-Ctf19∆C, Ctf19-Okp1 and Ctf19∆C-Okp1 fusion proteins from the rescue plasmid. Right panel: Western blot analysis visualizing the levels of the ectopically expressed, C-terminally 7xFLAG-tagged fusion proteins. Pgk1 levels are shown as loading control. (aa: Anchor-away) (B) Minichromosome loss assay. Chromosome segregation fidelity was determined in the Ctf19 anchor-away (SLI15/CTF19-FRB) strain, containing a minichromosome, either untransformed (-) or transformed with the indicated rescue constructs in the absence or presence of 1 µg/ml rapamycin. The percentage and standard error of red/red sectored colonies to the total colony number (white plus red/red sectored) of three biological replicates is shown. The results of 100% red colonies may be indicative of non-optimal conditions for the chromosome loss assay in combination with the anchor-away technique. (C) Localisation of ectopically expressed Ctf19-Okp1-GFP and Ctf19∆C-Okp1-GFP fusion proteins in the Ctf19 anchor-away strain (SLI15/CTF19-FRB) in the presence of 1 µg/ml rapamycin. Live cell fluorescence microscopy was performed 3 hr after rapamycin addition. Ndc80-mCherry was used as kinetochore marker. Merged mCherry and GFP signals are shown on the right. (BF: brightfield).
-
Figure 6—source data 1
Quantification of the minichromosome loss assay in a SLI15/CTF19-FRB strain.
- https://doi.org/10.7554/eLife.42879.012
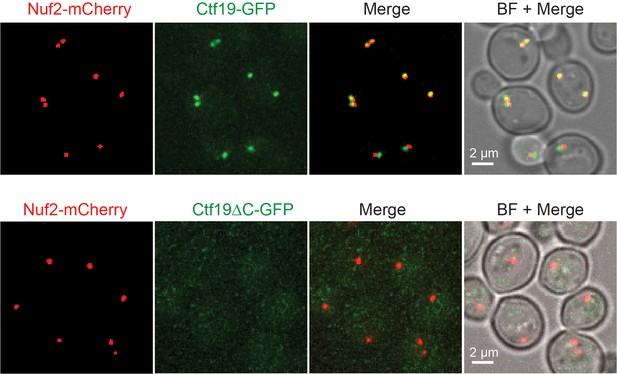
Ctf19∆C-GFP does not localize to kinetochores.
Live cell microscopy of non-synchronized cells grown in synthetic medium expressing either Ctf19-GFP or Ctf19∆C-GFP and Nuf2-mCherry as kinetochore marker. Merged mCherry and GFP signals are shown on the right.BF: Brightfield.
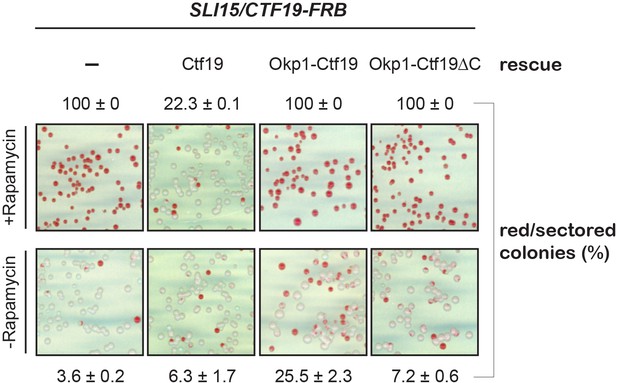
The N-terminal fusion protein of Ctf19 with Okp1 does not rescue chromosome segregation defects upon nuclear depletion of Ctf19 in the Sli15 wild-type background.
The loss of minichromosomes was assessed in the Ctf19 anchor-away (SLI15/CTF19-FRB) strain either untransformed (-) or transformed with the indicated rescue constructs in the absence or presence of 1 µg/ml rapamycin. The percentage and standard error of red/red sectored colonies to the total colony number (white plus red/red sectored) of three biological replicates is shown.
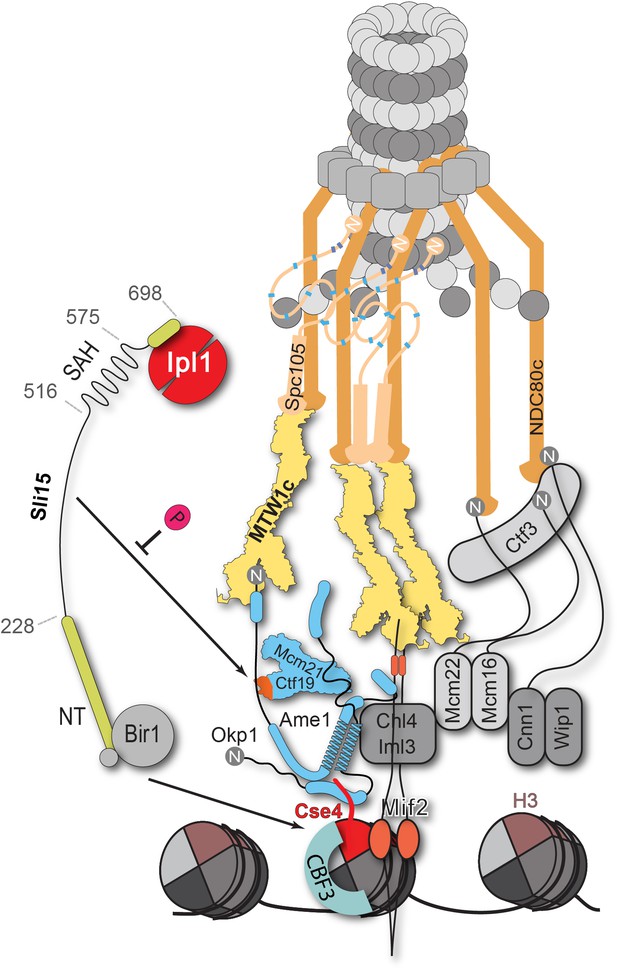
Schematic model of the budding yeast kinetochore subunit architecture.
The Okp1 core domain directly binds the essential motif of the Cse4 END suggesting that in contrast to humans, the dual recognition of Cse4-NCPs in S. cerevisiae is established by the essential inner kinetochore subunits Ame1/Okp1 and Mif2 through interaction with distinct Cse4 motifs. Together with the observation that Ctf19 associates with Sli15/Ipl1, further CPC interactions with the inner and outer kinetochore could be part of a kinetochore conformation that is dependent on Sli15INCENP. In line with the observed benomyl sensitivity of cells expressing Sli15ΔN as the only nuclear copy (Figure 5C), a recent study in Xenopus egg extracts found that CPC lacking the CEN domain of INCENP affected the correction of erroneous kinetochore-microtubule attachments (Haase et al., 2017). Centromere-targeting deficient CPC resulted in an imperfect inner kinetochore composition that failed to sense tension-loss and in intermediate Ndc80 phosphorylation levels that indicated the incapability of establishing a sharp phosphorylation gradient according to the spatial separation model. Flat Ndc80 phosphorylation levels could be sufficient for the non-selective turnover of erroneous kinetochore attachments, especially at budding yeast kinetochores which are attached to a single microtubule, unless cells are challenged by microtubule poisons.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (S. cerevisiae) | See Supplementary file 5 | |||
| Strain, strain background (S. cerevisiae) | S288c | |||
| Strain, strain background (E. coli) | BL21(DE3) | New England Biolabs | C2527 | |
| Strain, strain background (E. coli) | DH10Bac | ThermoFisher | 10361012 | |
| Cell line (S. frugiperda) | SF21; Spodoptera frugiperda | ThermoFisher | 11497013 | |
| Cell line (Trichoplusia ni) | High five; Trichoplusia ni | ThermoFisher | B85502 | |
| Genetic reagent (S. cerevisiae) | See Supplementary file 5 | |||
| Antibody | Anti-FLAG M2 (mouse monoclonal) | Sigma-Aldrich | F1804 RRID:AB_262044 | 1:5000 |
| Antibody | Anti-PGK1 (mouse monoclonal) | Invitrogen | 22C5D8 RRID:AB_2532235 | 1:10000 |
| Antibody | goat anti-mouse IgG-HRP | Santa Cruz Biotechnology | sc-2005 RRID:AB_631736 | 1:10000 |
| Recombinant DNA reagent | See Supplementary file 4 | |||
| Peptide, recombinant protein | 3xFLAG peptide | Ontores | ||
| Peptide, recombinant protein | lambda phosphatase | New England Biolabs | P0753S | |
| Commercial assay or kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs | E0552S | |
| Chemical compound, drug | BS3-H12/D12 cross-linker | Creative Molecules | 001SS | |
| Chemical compound, drug | Iodoacetamide | Sigma-Aldrich | I6125 | |
| Chemical compound, drug | Lysyl Endopeptidase | FUJIFILM Wako Pure Chemical Corporation | 125–05061 | |
| Chemical compound, drug | Trypsin Sequencing Grade Modified | Promega | V5111 | |
| Chemical compound, drug | SYBR Gold | ThermoFisher | S11494 | |
| Chemical compound, drug | AMP-PNP | Santa Cruz Biotechnology | CAS 72957-42-7 | |
| Chemical compound, drug | Rapamycin | Invitrogen | PHZ1235 | |
| Chemical compound, drug | Concanavalin A from Canavalia ensiformis | Sigma-Aldrich | C2010 | |
| Chemical compound, drug | FuGENE HD Transfection Reagent | Sigma-Aldrich | E2311 | |
| Chemical compound, drug | cOmplete ULTRA EDTA-free Protease Inhibitor Cocktail | Roche | 5892953001 | |
| Chemical compound, drug | Ni-NTA Agarose | Qiagen | 30210 | |
| Chemical compound, drug | Strep-Tactin Superflow Plus Agarose | Qiagen | 30004 | |
| Chemical compound, drug | M2 anti-FLAG agarose | Sigma-Aldrich | A4596 | |
| Other | Sep-Pak tC18 cartridges | Waters | WAT054960 | |
| Other | PD-10 Desalting Columns | GE Healthcare | 17085101 | |
| Other | µ-Slide 8 Well | Ibidi | 80826 | |
| Software, algorithm | xQuest | (Walzthoeni et al., 2012) | ||
| Software, algorithm | xVis | (Grimm et al., 2015) | ||
| Software, algorithm | Fiji | (Schindelin et al., 2012) | ||
| Software, algorithm | Clustal Omega | (Sievers et al., 2011) | ||
| Software, algorithm | SoftWoRx | GE Healthcare |
Additional files
-
Supplementary file 1
Inter- and intra-protein cross-links detected on in vitro reconstituted Cse4 containing nucleosomes interacting with the kinetochore complexes Ame1/Okp1, Ctf19/Mcm21, Mif2, Chl4/Iml3 and MTW1c.
- https://doi.org/10.7554/eLife.42879.014
-
Supplementary file 2
Inter- and intra-protein cross-links detected on in vitro reconstituted Sli15/Ipl1 interacting with the inner kinetochore proteins Ctf19, Okp1, Ame1 and Mcm21 (COMA).
- https://doi.org/10.7554/eLife.42879.015
-
Supplementary file 3
Predicted and experimentally annotated protein domains and motifs depicted in protein cross-link networks.
- https://doi.org/10.7554/eLife.42879.016
-
Supplementary file 4
Plasmids used in this study.
- https://doi.org/10.7554/eLife.42879.017
-
Supplementary file 5
Yeast strains used in this study.
- https://doi.org/10.7554/eLife.42879.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.42879.019





