Acetylation of BMAL1 by TIP60 controls BRD4-P-TEFb recruitment to circadian promoters
Figures
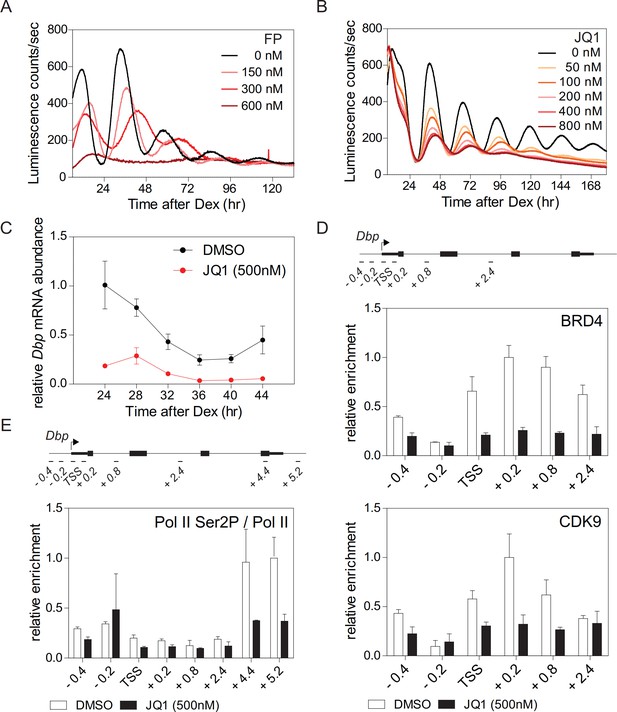
BRD4 controls clock gene expression.
(A and B) Bioluminescence recordings of dexamethasone (Dex) synchronized Bmal1-LUC fibroblasts treated with increasing concentrations of flavoperidol (A) or JQ1 (B) (n = 4). (C) Dbp mRNA dampening in Dex-synchronized fibroblasts by 500 nM JQ1 (n = 3; two-way ANOVA, see Supplementary file 1). (D and E) ChIP analysis of BRD4 and CDK9 (D) and Ser2-phosphorylated Pol II (normalized to total Pol II) (E) for the Dbp gene carried out 24 hr after Dex synchronization of the cells (n = 3). Schematic diagrams show the genomic structure of the Dbp locus. Horizontal lines represent PCR-amplified genomic regions. All data are shown as mean ± SD. Note that Dbp has several additional intronic Pol II pausing sites (Sobel et al., 2017). For validation of ChIP-grade quality of antibodies see Figure 1—figure supplement 2.
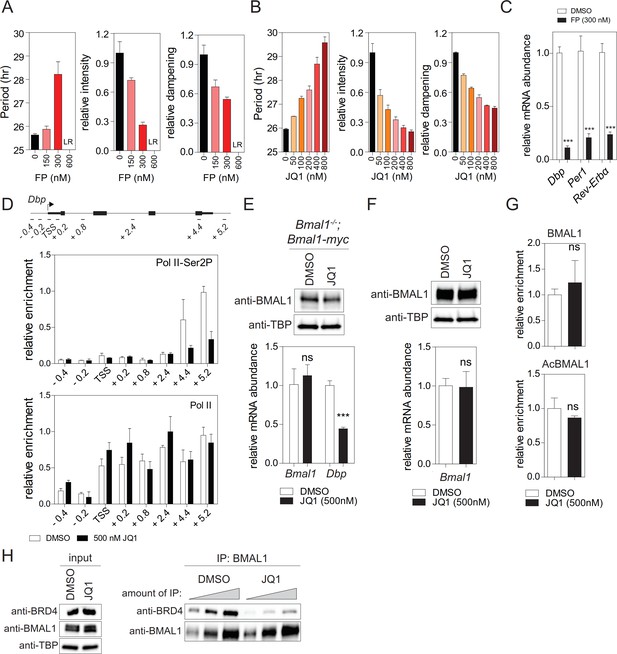
BRD4 controls expression of E-box-controlled clock genes.
(A - B) Change of period, luminescence intensity, and damping rate in flavopiridol (FP) (A) and JQ1 (B) treated dexamethasone (Dex) synchronized Bmal1-LUC fibroblasts (n = 4). LR: loss of rhythm. (C) mRNA expression analysis of fibroblasts treated with either DMSO or flavopiridol (300 nM) carried out 24 hr after Dex synchronization (n = 3). (D) Pol II and Ser2P-Pol II ChIP analysis of Dex synchronized DMSO or JQ1 treated fibroblasts (n = 3). (E) Immunoblot and mRNA expression analysis of Dex synchronized (24 hr after) Bmal1-/- MEFs transfected with empty vector or Bmal1-myc treated with either DMSO or JQ1 (500 nM) (n = 3). (F) BMAL1 protein abundance and Bmal1 mRNA expression in DMSO or JQ1 (500 nM) treated cells isolated 24 hr after Dex synchronization (n = 3). (G) ChIP analysis of BMAL1 and acetylated BMAL1 for the Dbp gene carried out 24 hr after Dex synchronization of MEFs (n = 3). (H) Interaction of BMAL1 with BRD4 in fibroblasts treated with either DMSO or JQ1 (500 nM). Immunoblots show results with increasing equivalents of IPs (two-fold dilutions) from nuclear extracts isolated from cells 24 hr after Dex synchronization. All data are shown as mean ± SD; ns > 0.05 and ***p<0.001 Student’s t-test).
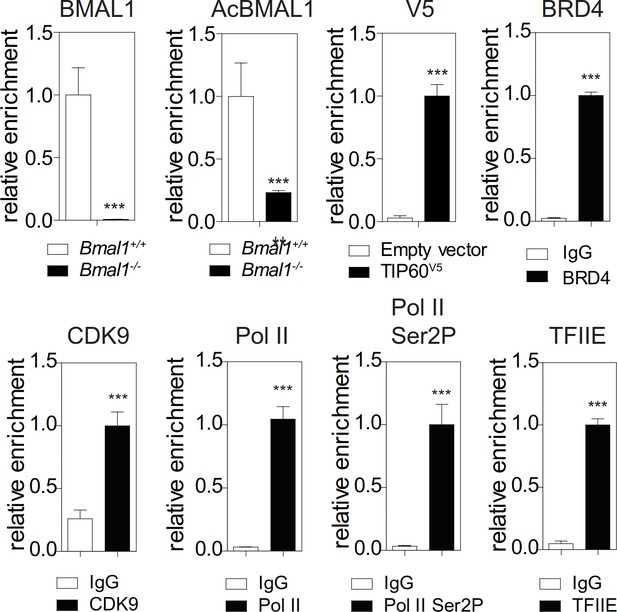
Validation of antibodies used for ChIP studies either deficient cells or antibody isotype controls were used to validate the ChIP-grade quality of the antisera.
The test gene used was Dbp. Data are shown as mean ± SD (n = 3; **p<0.01, and ***p<0.001 Student’s t-test).
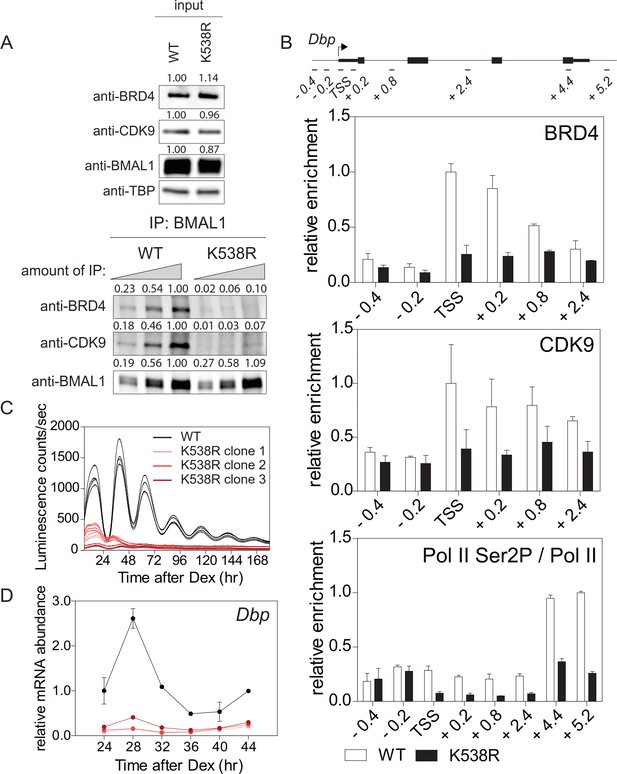
Lys538 acetylation of BMAL1 Is essential for transcription elongation.
(A) Interaction of BMAL1 or BMAL1K538R with BRD4 and CDK9 as shown by immunoblotting of nuclear extracts from synchronized fibroblast 24 hr after Dex treatment (two-fold dilutions). TBP (TATA-binding protein) was used as loading control. Numerical values represent intensities of chemiluminescence signals of individual bands, normalized to wildtype and loading control for input samples. (B) ChIP analysis of BRD4, CDK9 and Ser2P-Pol II (normalized to total Pol II) binding to the Dbp gene in wildtype and BMAL1K538R synchronized fibroblasts as shown in (A) (n = 3). (C) Bmal1-LUC bioluminescence tracings of synchronized wildtype fibroblasts and of three independent BMAL1K538R clones (n = 4). (D) Dbp mRNA expression analysis of wildtype or BMAL1K538R fibroblasts taken from the experiment shown in (C) (n = 3, two-way ANOVA, see Supplementary file 1). All data are shown as mean ± SD.
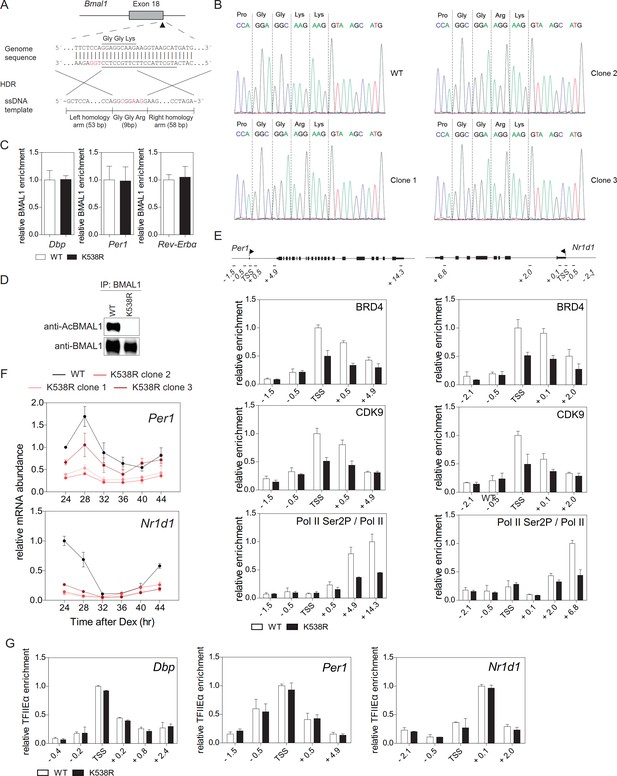
CRISPR/Cas9-mediated generation and characterization of BMAL1K538Rmutant cells.
(A) Schematic representation of the gene targeting strategy for generating BMAL1K538R mutant cells using the CRISPR/Cas9 system. The gRNA-targeting sequence is underlined and the PAM sequence is indicated in red. The oligonucleotide donor (120 bp) is shown below the targeted site, with the Lys-to-Arg substitution indicated in red. Silent point mutations were introduced for the two adjacent Gly to prevent cutting of the donor DNA or re-cutting of the genome after homology-directed recombination (shown in red). (B) Representative sequencing results from PCR amplicons of the Lys538 region from three individual clones that were subsequently used for experimental work. Primer sequences are specified in Supplementary file 2. (C) ChIP analysis of BMAL1 binding to Dbp, Per1, and Nr1d1 genes in wildtype and BMAL1K538R synchronized fibroblasts 24 hr after Dex synchronization (n = 3). (D) BMAL1 IPs from wildtype or BMAL1K538R fibroblast nuclear extracts carried out 24 hr after Dex synchronization of fibroblasts were immunoblotted with anti-AcBMAL1 and anti-BMAL1 antibodies. (E) BRD4, CDK9, and Ser2P-Pol II (normalized to total Pol II) ChIP analysis of Dex synchronized wildtype or BMAL1K538R fibroblasts (n = 3). (F) Per1 and Nr1d1 mRNA expression analysis of wildtype or BMAL1K538R fibroblasts (n = 3, two-way ANOVA, see Supplementary file 1). (G) ChIP analysis of TFIIEα binding to Dbp, Per1, and Nr1d1 genes in wildtype and BMAL1K538R synchronized fibroblasts (n = 3). All data are shown as mean ± SD.
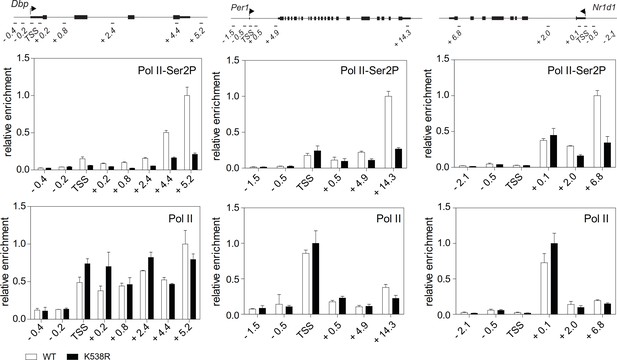
Characterization of BMAL1K538Rmutant cells Pol II and Ser2P-Pol II ChIP analysis of Dex synchronized wildtype or BMAL1K538R fibroblasts (n = 3).
All data are shown as mean ± SD.
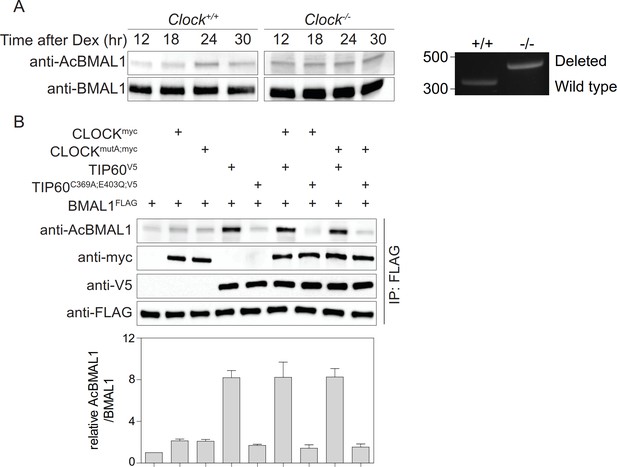
Acetylation of BMAL1 in CLOCK-deficient cells.
(A) BMAL1 IP analysis of control and Clock-deficient MEF nuclear extracts over a 24 hr time course. For both genotypes, equivalent amounts of BMAL1 were included as shown in the anti-BMAL1 probed western blot. Lys538 acetylation of BMAL1 was detected using an anti-AcBMAL1 antibody (left). Wildtype and mutant cells were PCR-genotyped to confirm their genotype (right) (Debruyne et al., 2006). (B) BMAL1FLAG, CLOCKmyc, CLOCKmutA;myc, TIP60V5, and TIP60C369A;E403Q;V5 were transiently overexpressed in HEK293T cells in the combinations indicated. Lysates were subjected to IPs and immunoblotted with antibodies indicated (n = 3). Data are shown as mean ± SD.
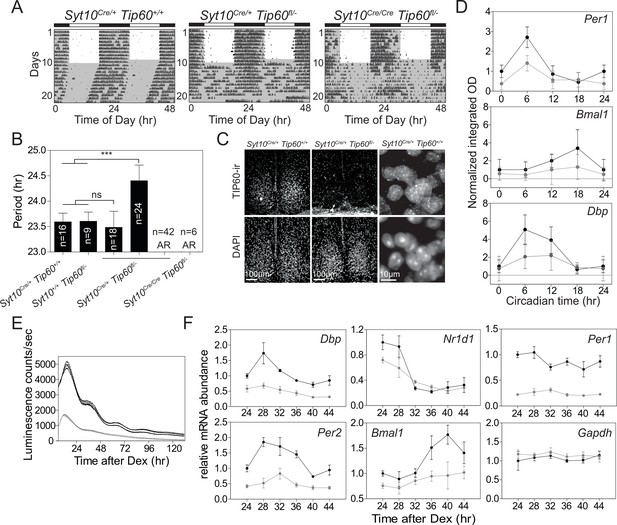
TIP60-deficiency evokes a circadian phenotype in mice and disrupts rhythmic clock gene expression in the SCN and in MEFs.
(A) Double-plotted actograms of control (Syt10Cre/+Tip60+/+) and mutant (Syt10Cre/+Tip60 fl/-, Syt10Cre/Cre Tip60fl/-) mice under 12 hr:12 hr light-dark and constant darkness conditions. Gray shadings indicate dark phases. (B) Free-running periods determined by χ2 periodogram analysis. AR: arrhythmic (***p<0.001, one-way ANOVA with Bonferroni post-test). (C) TIP60-immunoreactivity (ir) in the SCN of control (Syt10Cre/+Tip60+/+) and arrhythmic mutant (Syt10Cre/+Tip60 fl/-) mice. (D) Densitometric quantification of clock gene mRNA expression from radioactive in situ hybridization analysis of control (Syt10Cre/+Tip60+/+; black) and mutant (Syt10Cre/+Tip60 fl/-; gray) SCN sections (n = 3; two-way ANOVA, see Supplementary file 1). (E) Bioluminescence recordings of synchronized Tip60fl/-; Bmal1-LUC MEFs transduced with AdGFP (black) or AdCre (gray) (n = 4). (F) mRNA expression analysis of Dex synchronized Tip60fl/- MEFs transduced with either AdGFP (black) or AdCre (gray) (n = 3; two-way ANOVA, see Supplementary file 1). All data are shown as mean ± SD.
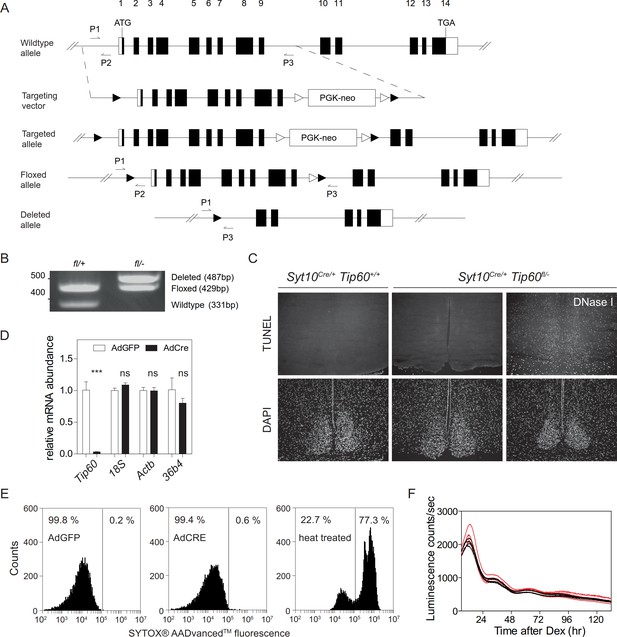
Generation and validation of the Tip60-deficient mice and MEFs derived from such animals.
(A) Targeting strategy for the Tip60 locus. Tip60 wildtype allele, targeting vector, targeted allele, floxed allele after removal of the PGK-Neo cassette, and the mutant Tip60 allele created by Cre-mediated recombination are shown. LoxP (black triangle) and FRT (open triangle) sites are marked. The arrows indicate the position of diagnostic primers P1-P3 (see Supplementary file 1). (B) PCR genotyping of Tip60 alleles. The wildtype allele produces a 331 bp amplicon (primers P1, (P2) while the floxed allele produces a 429 bp amplicon (P1, P2). Deletion of the LoxP-flanked region of the Tip60 locus leads to a 487 bp fragment (P1, P3). (C) TUNEL immunoreactivity on brain sections of Syt10Cre/+Tip60+/+ (control), arrhythmic Syt10Cre/+Tip60 fl/- animals, and Syt10Cre/+Tip60 fl/- sections treated in situ with DNase I. (D) mRNA expression of housekeeping genes in Tip60fl/- MEFs transduced with either AdGFP or AdCre, 24 hr after Dex synchronization. Data are mean ± SD (n = 3; ns > 0.05 and ***p<0.001, Student’s t-test). (E) Flow cytometric analysis of AdGFP, AdCre transduced, and heat-treated (3 min, 95°C) confluent Tip60fl/- MEFs. (F) Bioluminescence tracing of Dex-synchronized Bmal1-LUC MEFs transduced with AdGFP (black) or AdCre (red) (n = 4).
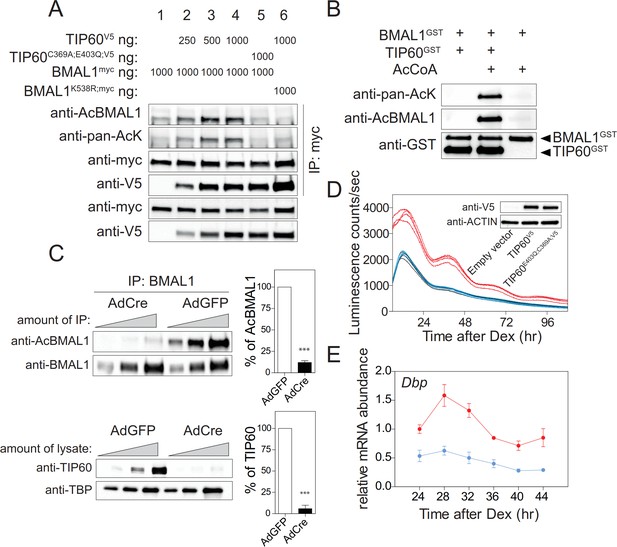
TIP60 acetylates BMAL1.
(A) BMAL1myc, BMAL1K538R, TIP60V5, and TIP60C369A;E403Q;V5 were transiently overexpressed in HEK293T cells in the combinations indicated on top. Lysates were subjected to IPs using the antibodies indicated. (B) Recombinant BMAL1GST, TIP60GST and acetyl-CoA were incubated and the presence of acetylated BMAL1 was detected by immunoblotting using either a pan-acetyl or a Lys538-specific AcBMAL1 antibody. (C) IPs (top) and nuclear extracts (bottom) from AdGFP (control) and AdCre (mutant) transduced unsynchronized Tip60fl/- MEF (two-fold dilutions) were immunoblotted with antibodies indicated and signal intensities were quantified and normalized. Data are shown as mean of all relative values ± SD (n = 3). (D) Tip60fl/-; Bmal1-LUC MEFs stably expressing TIP60V5 (red), TIP60C369A;E403Q;V5 (blue) or empty vector (black, coinciding with the blue tracing) were transduced with AdCre and bioluminescence was recorded (n = 4). (E) Dbp expression profiles in TIP60V5 and TIP60C369A;E403Q;V5 cells shown in (D). (n = 3; two-way ANOVA, see Supplementary file 1).
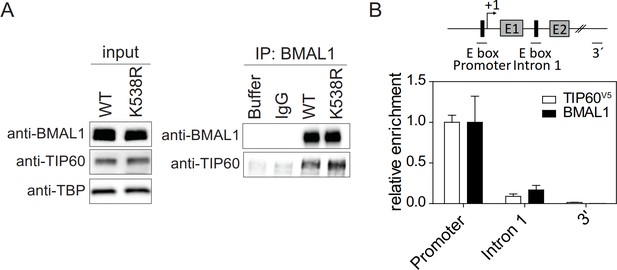
TIP60 interacts with BMAL1 on chromatin.
(A) Immunoblot analysis of the interaction between endogenous TIP60 with BMAL1 or BMAL1K538R. IPs from Dex synchronized MEF (24 hr after Dex) nuclear extracts were immunoblotted with the antibodies indicated. IgG and buffer (mock IP) served as negative controls. (B) ChIP analysis of Tip60-/-; Tip60V5 MEFs using anti-V5 and anti-BMAL1 antibodies. The diagram shows the 5’ region of the Dbp gene. E-boxes that bind CLOCK-BMAL1 at exons 1 (E1), exon 2 (E2), and transcription start sites are shown. Horizontal lines represent PCR-amplified genomic regions. The 3’ UTR region was used as a control. Data are shown as mean ± SD (n = 3).
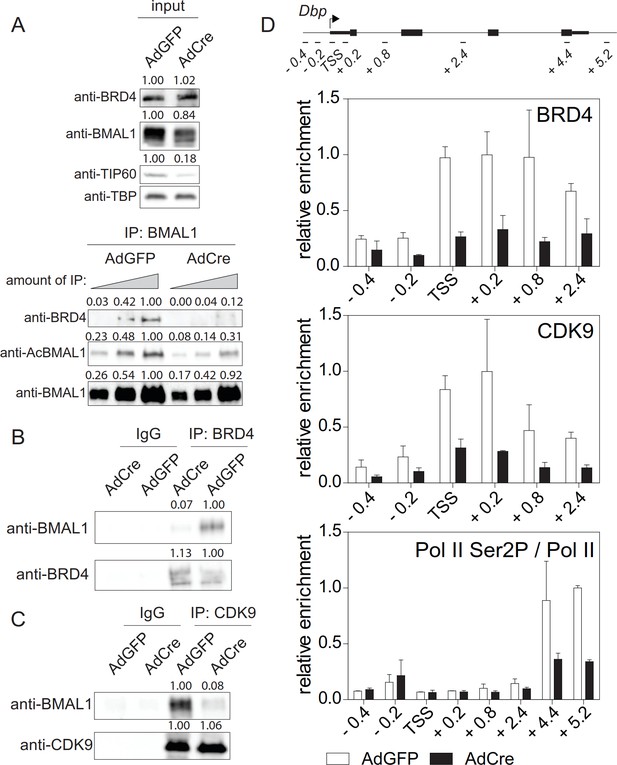
TIP60 controls productive elongation.
(A) Interaction of BMAL1 with BRD4 in Tip60fl/- MEFs transduced with either AdGFP (control) or AdCre (mutant). Immunoblots show results with increasing equivalents of IPs (two-fold dilutions) from nuclear extracts isolated from cells 24 hr after Dex synchronization. (B) BRD4 or (C) CDK9 IPs from AdGFP or AdCre transduced Tip60fl/- MEF nuclear extracts were immunoblotted with antibodies indicated. Analyses were carried out 24 hr after Dex synchronization. (D) ChIP analysis for BRD4, CDK9 and Ser2P-Pol II (normalized to total Pol II) in the promoter or the 3’-end of Dbp for the cells shown in (A). Data are shown as mean ± SD (n = 3). Numerical values represent intensities of chemiluminescence signals of individual bands, normalized to wildtype and loading control for input samples.
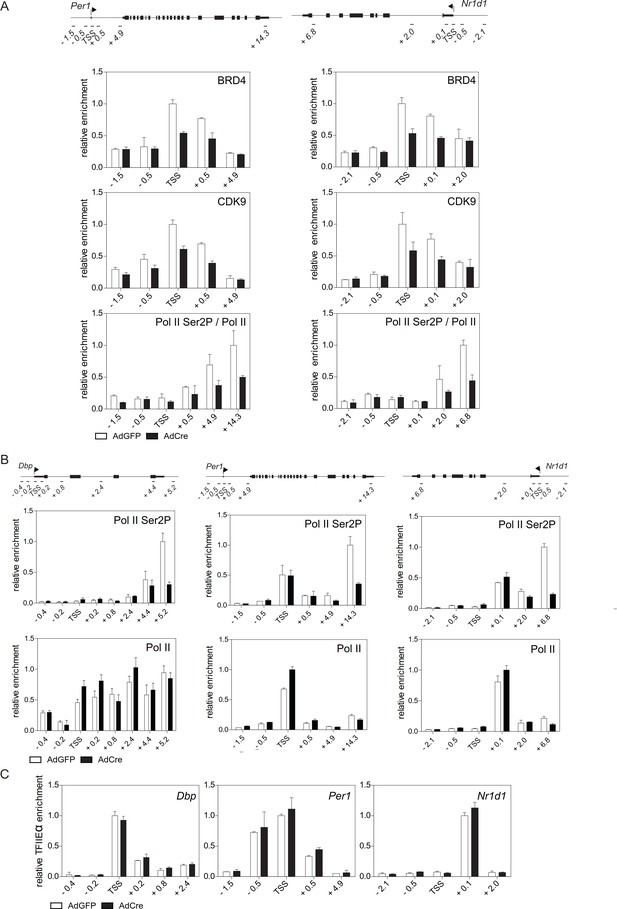
TIP60 controls productive elongation of circadian genes.
(A, B and C) BRD4, CDK9, and Ser2P-Pol II (normalized to total Pol II) (A), Pol two and Ser2P-Pol II (B) or TFIIEα (C) ChIP analysis of Dex synchronized Tip60fl/- MEFs transduced with either AdGFP or AdCre. All data are shown as mean ± SD (n = 3).
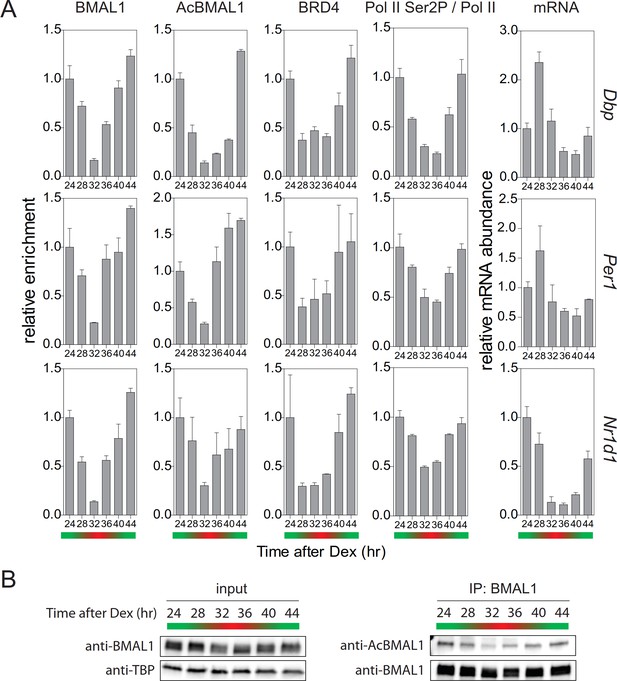
Rhythmic profile of productive elongation.
(A) ChIP profiles of Dex-synchronized MEFs over a course of 24 hr. ChIP shows time-of-day-dependent enrichment of BMAL1, acetylated BMAL1, BRD4, and Ser2P-Pol II (normalized to total Pol II) at the promoter or 3’-end (Ser2P-Pol II) of Dbp, Per1, and Nr1d1 genes. The rightmost column reveals time-dependence of mRNA accumulation for these genes. Data are shown as mean ± SD (n = 3). (B) Immunoblot analysis with the indicated antibodies of Dex-synchronized fibroblast nuclear extracts and BMAL1 IPs. The color bars represent the activation (green) and repression (red) phases of the circadian cycle.
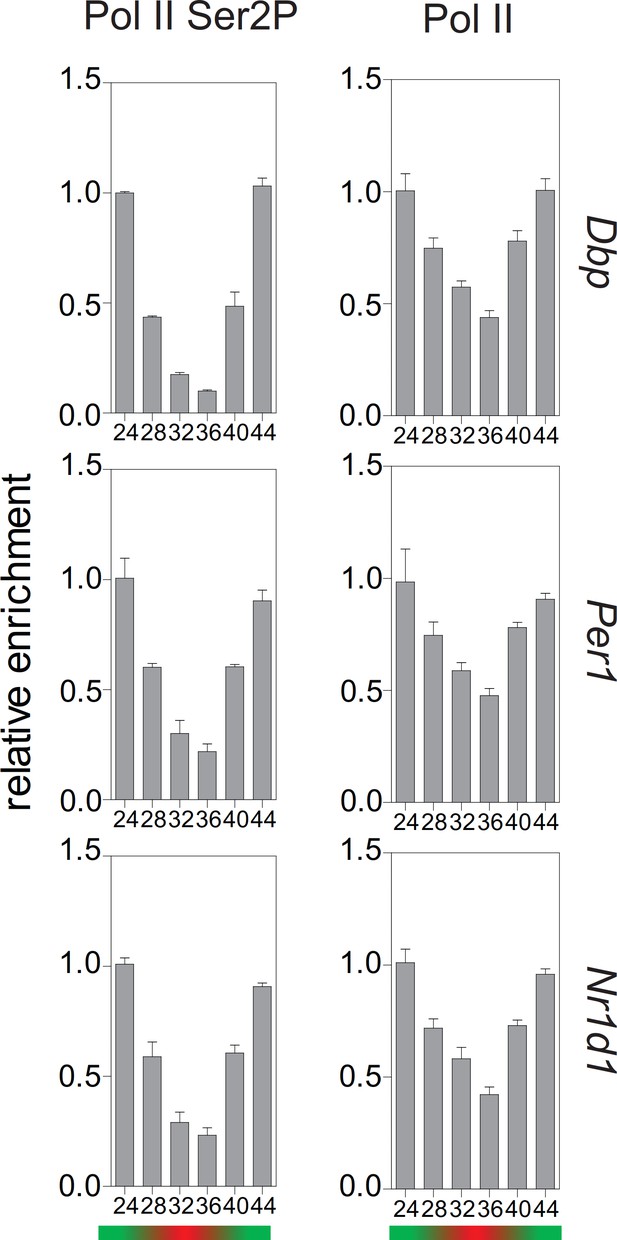
Rhythmic profile of Pol II and Ser2P-Pol II abundance ChIP profiles of Dex-synchronized MEFs over a course of 24 hr.
ChIP shows time-of-day-dependent enrichment of Pol II and Ser2P-Pol II at the 3’-end of Dbp, Per1, and Nr1d1 genes. Data are shown as mean ± SD (n = 3).
Additional files
-
Supplementary file 1
Two-way ANOVA statistical analysis.
- https://doi.org/10.7554/eLife.43235.017
-
Supplementary file 2
Primer sequences.
- https://doi.org/10.7554/eLife.43235.018
-
Supplementary file 3
Key resources table.
- https://doi.org/10.7554/eLife.43235.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43235.020





