Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPARγ
Figures
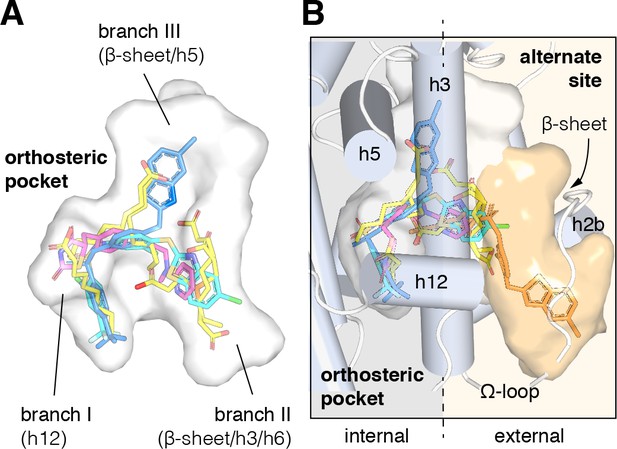
Structural locations of PPARγ orthosteric pocket and alternate site.
(A) The T/Y-shaped orthosteric pocket can accommodate one or more natural ligands such as nonanoic acid (C9; yellow sticks; PDB: 4EM9) and synthetic ligands such as rosiglitazone (pink sticks; PDB: 4EMA) or T2384 (light and dark blue sticks representing different crystallized binding modes; PDB: 3K8S, chain A and B, respectively). (B) The orthosteric pocket (white pocket surface) is completely enclosed within the alpha helical sandwich fold of the ligand-binding domain (LBD). Ligands such as T2384 (orange sticks; PDB: 3K8S, chain B) can also bind to a solvent accessible alternate site (orange pocket surface) distinct from the orthosteric pocket, structurally defined as the region between helix 3 (h3) and the flexible Ω-loop (dotted line separating the region internal to the LBD, grey background; and external to the LBD, light orange background).
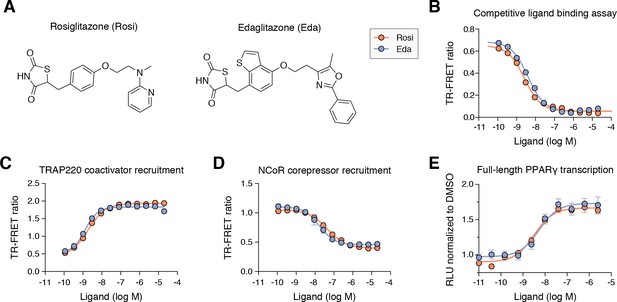
Edaglitazone is a thiazolidinedione (TZD) PPARγ agonist related to rosiglitazone.
(A) Chemical structures of edaglitazone and rosiglitazone. (B) TR-FRET ligand displacement assay. Data plotted as the average and S.D. of three experimental replicates. (C,D) TR-FRET coregulator interaction assay data in the presence of peptides derived from (C) TRAP220 coactivator and (D) NCoR corepressor. Data plotted as the average and S.D. of three experimental replicates. (E) Cell-based luciferase reporter assay measuring full-length PPARγ transcription in HEK293T cells; RLU = relative luciferase units. Data plotted as the average and S.E.M. of four experimental replicates.
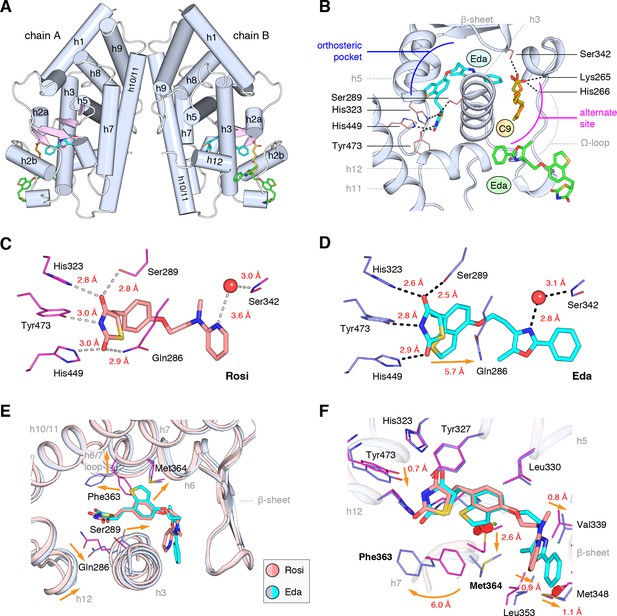
Co-crystal structure of edaglitazone-bound PPARγ LBD reveals a cobound bacterial MCFA.
(A) Overall structure (helices, light blue; strands, pink) with two bound edaglitazone (EDA) ligands, one to the canonical orthosteric pocket (blue) and another to a surface pocket (green), and a C9 ligand bound to an alternate site (yellow and orange in chain A and B, respectively). (B) Molecular interactions between PPARγ and the EDA and C9 ligands. Dotted lines denote hydrogen bonds. (C,D) Comparison of the hydrogen bond interactions between PPARγ and (C) rosiglitazone (PDB: 2PRG) and (D) edaglitazone. Dotted lines denote hydrogen bonds. (E,F) Conformational changes relative to rosiglitazone-bound PPARγ (PDB: 2PRG) that allow the orthosteric pocket to adapt to binding the bulkier benzo[b]thiophene moiety in edaglitazone. The changes (orange arrows) include (E) shifting of the backbone of helix 3, helix 6, helix 6/7 loop, and helix 12; and (F) large movements of the Phe363 and M364 side chains adjacent to the benzo[b]thiophene moiety. Also see Figure 3—figure supplements 1 and 2.
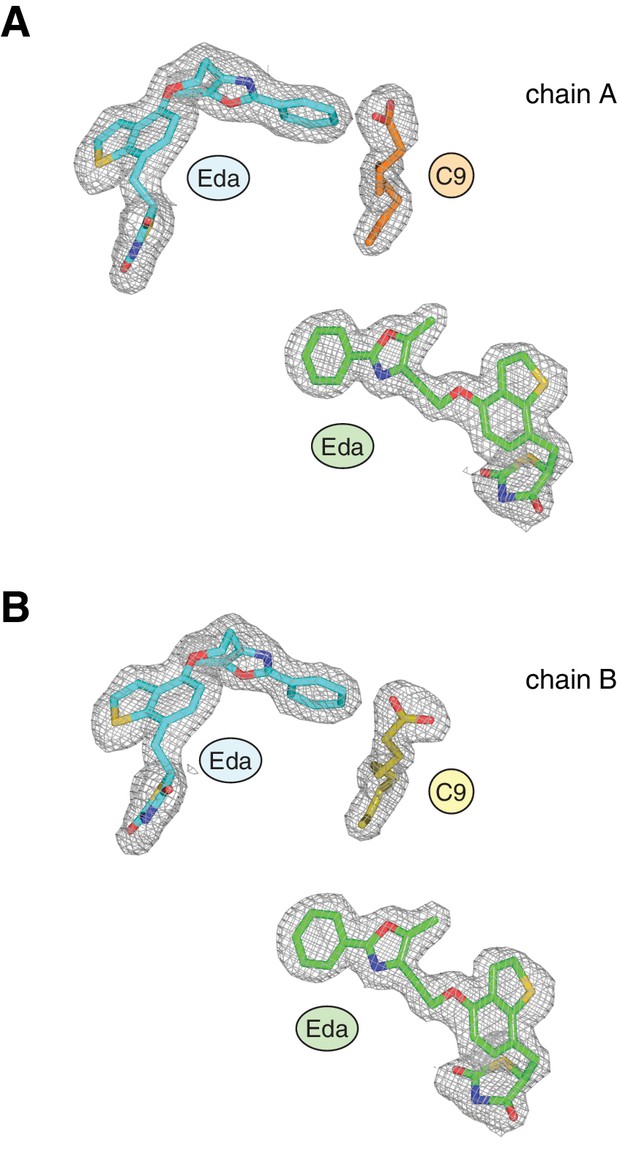
Omit map (2FO–FC, contoured at 1 σ) of the EDA and C9 ligands in the crystal structure.
Ligands in chains A and B are shown in (A) and (B), respectively, and colored as follows: EDA bound to the canonical orthosteric pocket (blue), EDA bound to a surface pocket (green), and C9 bound to the alternate site (yellow and orange in chain A and B, respectively). Figure 3—figure supplement 2. The TZD head group of the second edaglitazone molecule docks into the PPARγ AF-2 surface (A,B) in a similar manner to a SRC-1 coactivator peptide bound to the AF-2 surface (C) in the crystal structure of rosiglitazone-bound PPARγ (PDB: 2PRG). The hydrophobic tail group of this second edaglitazone molecule packs within a surface pocket created by the Ω-loop and the loop connecting helix 11 and 12 (B). The second EDA ligand bridges interactions between two PPARγ molecules in different unit cells within the crystal lattice by binding to the Ω-loop region in one molecule (e.g., chain A) and the AF-2 surface of a symmetry related molecule (e.g., chain B’).
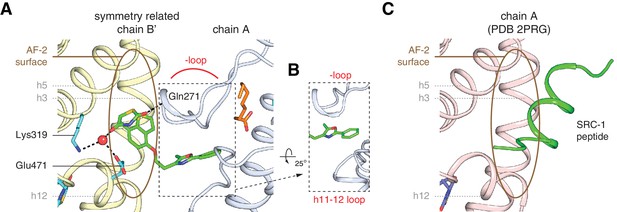
The TZD head group of the second edaglitazone molecule docks into the PPARγ AF-2 surface.
(A,B) in a similar manner to a SRC-1 coactivator peptide bound to the AF-2 surface (C) in the crystal structure of rosiglitazone-bound PPARγ (PDB: 2PRG). The hydrophobic tail group of this second edaglitazone molecule packs within a surface pocket created by the Ω-loop and the loop connecting helix 11 and 12 (B). The second EDA ligand bridges interactions between two PPARγ molecules in different unit cells within the crystal lattice by binding to the Ω-loop region in one molecule (e.g., chain A) and the AF-2 surface of a symmetry related molecule (e.g., chain B’).
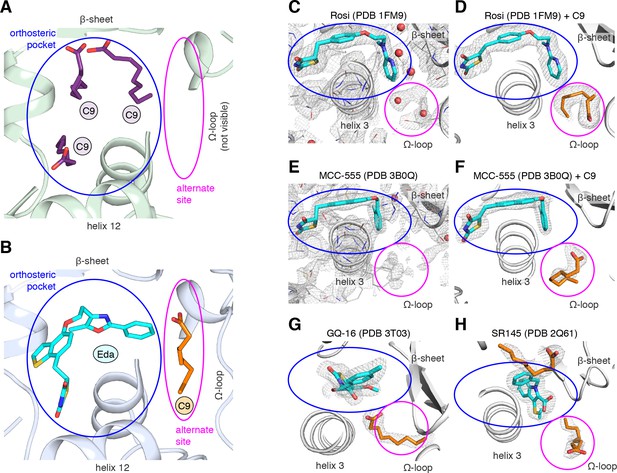
MCFA electron density in previously solved synthetic ligand-bound PPARγ LBD crystal structures.
(A,B) Comparison of a crystal structure of PPARγ LBD with (A) three molecule of C9 bound to the orthosteric pocket (PDB: 4EM9) and (B) C9 bound to the alternate site with edaglitazone bound to the orthosteric pocket. (C–F) Electron density maps for previously determined TZD-bound (blue) crystal structures shown with (C,E) original deposited structural models and (D,F) C9 (orange) modeled and refined into density present in the alternate site. (G,H) C9 (orange) modeled and refined into previously determined crystal structures of PPARγ bound to (G) GQ-16 (blue) and (H) SR145 (blue). Omit maps (2FO–FC) contoured at 1 σ in all but the C9 modeled into the GQ-16 structure, which was contoured at 0.5 σ. The orthosteric pocket and alternate site are circled in blue and magenta, respectively.
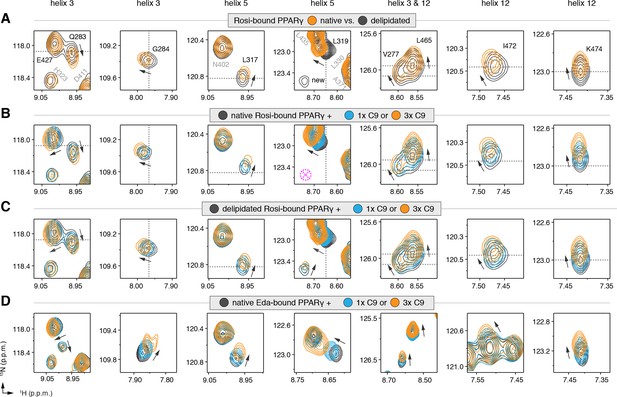
NMR confirms cobinding of exogenously added C9 and synthetic TZDs.
Differential 2D [1H,15N]-TROSY-HSQC NMR data comparing (A) native vs. delipidated PPARγ LBD bound to rosiglitazone (two equiv.); (B) C9 added into native PPARγ LBD bound to rosiglitazone (two equiv.); (C) C9 added into delipidated PPARγ LBD bound to rosiglitazone (two equiv.); and (D) C9 added into native PPARγ LBD bound to edaglitazone (two equiv.). The dotted lines in panels A–C represent the vertical (1H chemical shift) or horizontal (15N chemical shift) position of the peaks in the delipidated form to illustrate how the peaks corresponding to the native, nondelipidated form shift ‘on path’ to changes caused by C9 binding. Black arrows denote the chemical shift changes; pink dotted circle/cross denotes a missing peak; and dotted lines denote the delipidated apo-PPARγ peak positions to illustrate the ‘on path’ transitions of native and C9-bound forms. Also see Figure 5—figure supplement 1–5.
Full differential 2D [1H,15N]-TROSY-HSQC NMR data comparing 15N-PPARγ LBD in the native form (orange) vs. delipidated using a refolding procedure (black) or treatment using LIPIDEX 1000 resin (green), the latter two of which are nearly identical and show an increase in the intensities of some peaks vs. native.
https://doi.org/10.7554/eLife.43320.010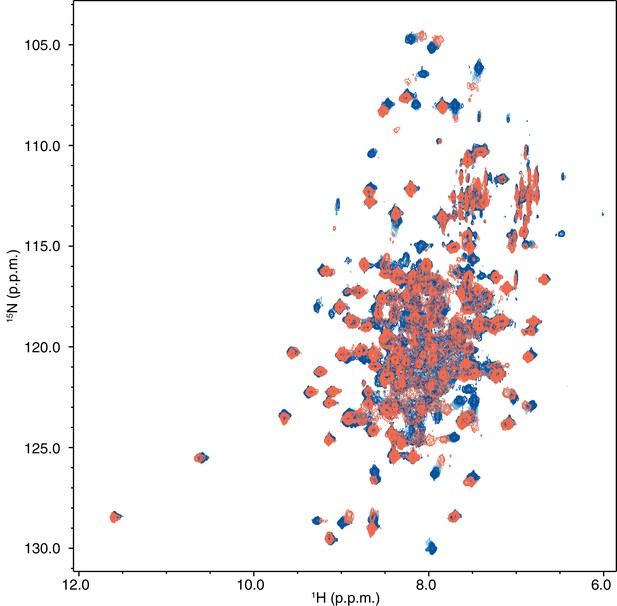
Full differential 2D [1H,15N]-TROSY-HSQC NMR data comparing C9 (nonanoic acid; 1, 2, 3, and four equiv.; light to dark blue) added into native 15N-PPARγ LBD (red).
https://doi.org/10.7554/eLife.43320.011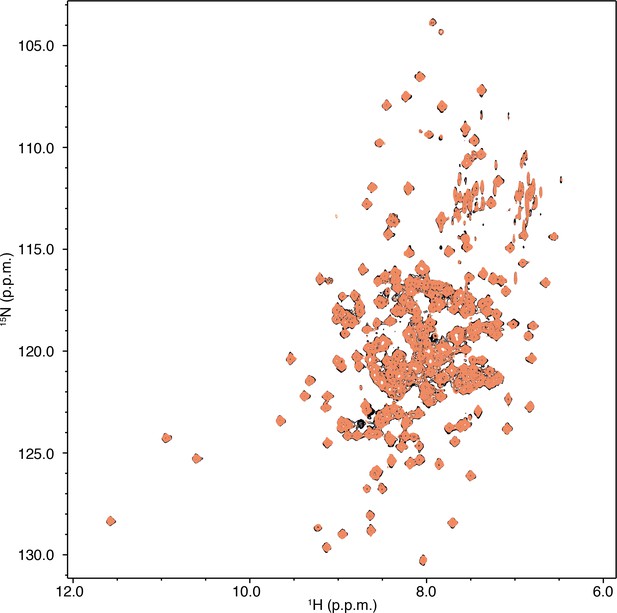
Full differential 2D [1H,15N]-TROSY-HSQC NMR data comparing native (red) vs.delipidated (black) PPARγ bound to rosiglitazone (two equiv.).
https://doi.org/10.7554/eLife.43320.012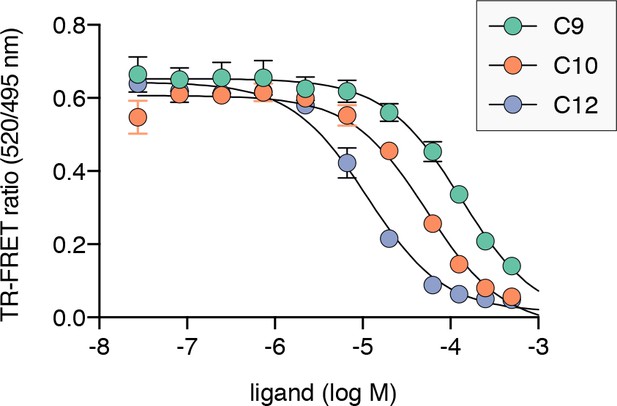
TR-FRET based PPARγ LBD ligand displacement assay for nonanoic acid (C9; Ki = 47.4 μM), decanoic acid (C10; 19.4 μM), and dodecanoic acid (C12; 4.1 μM).
Data plotted as the average and S.D. of three experimental replicates.
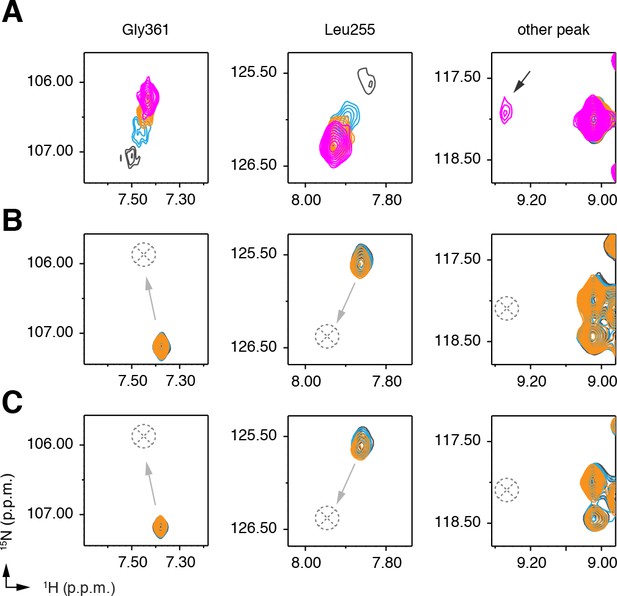
Differential 2D [1H,15N]-TROSY-HSQC NMR data showing that chemical shifts corresponding to residues (G361, L255, and an unassigned peak) perturbed when.
(A) C9 (1, 2, and three equiv. ; blue, orange, and magenta, respectively) binds to native PPARγ (black), (B,C) but not when C9 (1 and 3 equiv.; blue and orange, respectively) binds to rosiglitazone-bound (two equiv.) PPARγ in the (B) delipidated or (C) native form. The black arrow denotes a peak present when PPARγ is bound to C9; grey arrows denote the lack of transition to a C9-bound peak, which is missing in the rosiglitazone-bound data (dotted circle/cross).
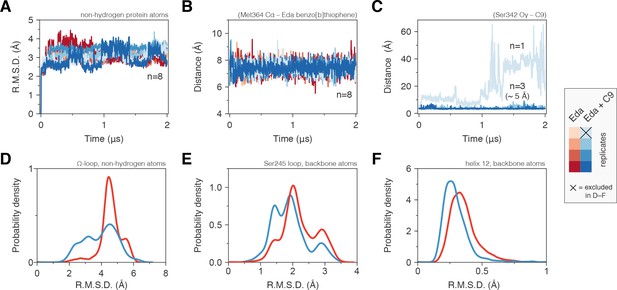
Molecular simulations reveal C9 cobinding stabilizes the PPARγ LBD.
(Α) Heavy atom R.M.S.D. to the starting conformation (crystal structure after minimization and equilibration) reveal that all four simulations bound to edaglitazone only to the orthosteric site (red lines) or cobound to C9 at the alternate site (blue lines) are stable during the simulation period. (B) Distance between benzo[b]thiophene group in edaglitazone and the M364 Cα atom shows that edaglitazone remains stably bound to the orthosteric pocket in all eight simulations. (C) C9 remains stably bound to the alternate site in three of four cobound simulations. In the legend, the × denotes the Eda + C9 (light blue) simulation where C9 unbound during the simulation, which was excluded from the analysis. (D–F) Probability density histogram distributions of the R.M.S.D. relative to the starting structure for (D) the Ω-loop, (E) the loop containing the serine residue phosphorylated by Cdk5, and (F) helix 12. Also see Figure 6—figure supplement 1.
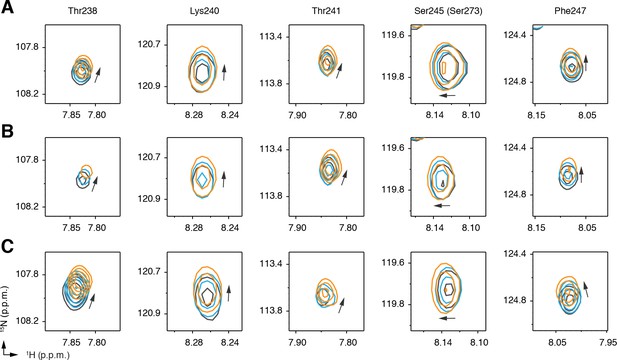
Differential 2D [1H,15N]-TROSY-HSQC NMR data focusing on residues in the Ser273 loop comparing C9 (1 and 3 equiv.; blue and orange, respectively) added into.
(A) delipidated PPARγ bound to rosiglitazone (two equiv.; black); (B) native PPARγ bound to rosiglitazone (two equiv.; black); and (C) native PPARγ bound to edaglitazone (two equiv.; black).
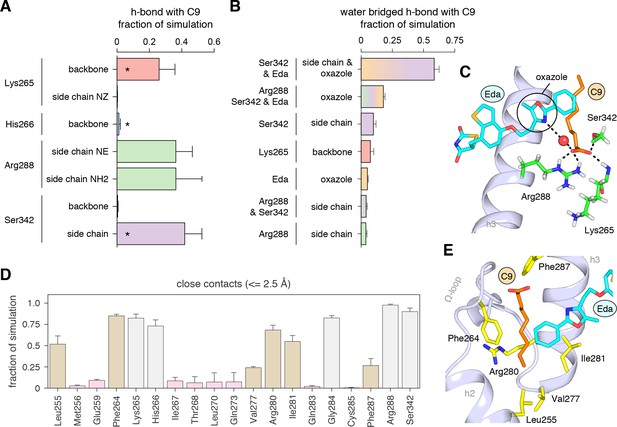
Interactions driving cobinding of C9 to the alternate site.
(A) Direct and (B) water-bridged hydrogen bonds detected in the three 2 μs molecular simulations where C9 remained stably bound; * denotes hydrogen bonds observed in crystal structure. Data plotted as the average and S.D. of three experimental replicates. (C) Example of the C9-mediated hydrogen bond network from a snapshot of the simulation trajectory. (D) Close atomic contacts populated in the simulation (brown, hydrophobic side chain contacts; grey, other contacts; pink, lower abundance contacts). Data plotted as the average and S.D. of three experimental replicates. (E) Structural example of the hydrophobic contacts (brown in D).
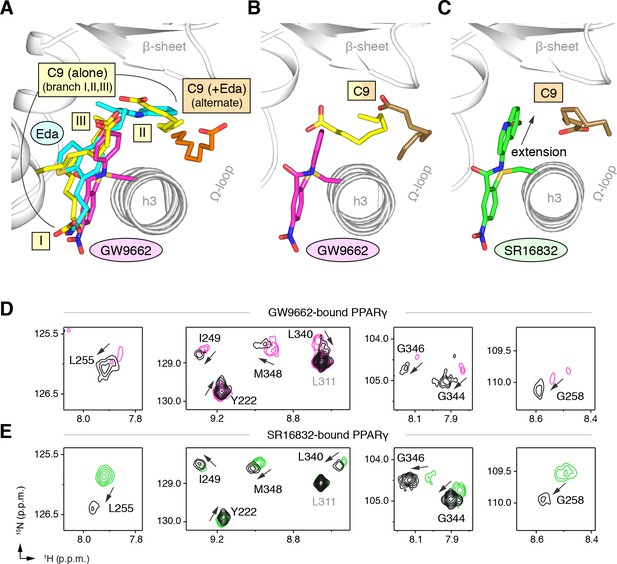
Covalent orthosteric antagonists isolate C9 cobinding.
(A) The covalent antagonist GW9662 (PDB: 3B0R; magenta) overlaps with the crystallized binding modes of 2 out of 3 C9 molecules bound to the orthosteric pocket (PDB: 4EM9) but not C9 bound to the alternate site. (B,C) Crystal structures of PPARγ LBD bound to covalent antagonists (B) GW9662 and (C) SR16832 cobound to C9. (D,E) Differential 2D [1H,15N]-TROSY-HSQC NMR data of PPARγ LBD covalently bound to (D) GW9662 (magenta) or (E) SR16832 (green) confirms cobinding of C9 (black; two equiv.); arrows denote chemical shift changes. Also see Figure 8—figure supplement 1 and 2.
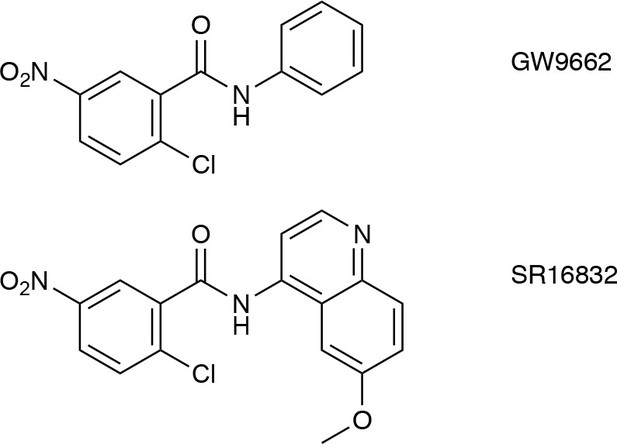
Chemical structures of covalent orthosteric ligands used in this study.
https://doi.org/10.7554/eLife.43320.019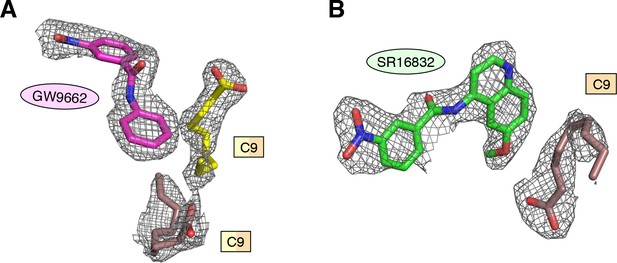
Omit map (2FO–FC, contoured at 1 σ) of the ligands in the (A) GW9662-bound and (B) SR16832-bound crystal structures of PPARγ LBD with cobound bacterial C9 ligands.
https://doi.org/10.7554/eLife.43320.020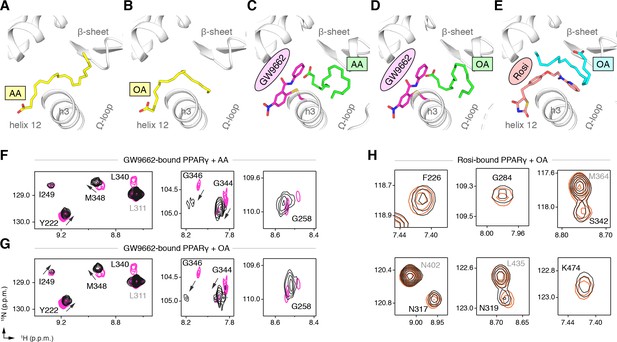
Cobinding of endogenous UFAs and synthetic PPARγ ligands.
(A,B) UFA ligand binding poses in crystal structures of PPARγ LBD (chain A) bound to (A) AA and (B) OA. (C–E) Ligand binding poses in crystal structures obtained from UFA-bound co-crystals soaked with synthetic ligands, including (C) AA-bound PPARγ LBD soaked with GW9662, (D) OA-bound PPARγ LBD soaked with GW9662, and (E) OA-bound PPARγ LBD soaked with rosiglitazone. (F,G) Differential 2D [1H,15N]-TROSY-HSQC NMR data of PPARγ LBD covalently bound to GW9662 (magenta) confirming cobinding with (F) AA (black; one equiv.). and (G) OA (black; one equiv.). (H) Differential 2D [1H,15N]-TROSY-HSQC NMR data of PPARγ LBD bound to rosiglitazone (pink; two equiv.) confirming cobinding with OA (black; one equiv.). Black arrows denote the chemical shift changes in the NMR data. Also see Figure 9—figure supplements 1 and 2.
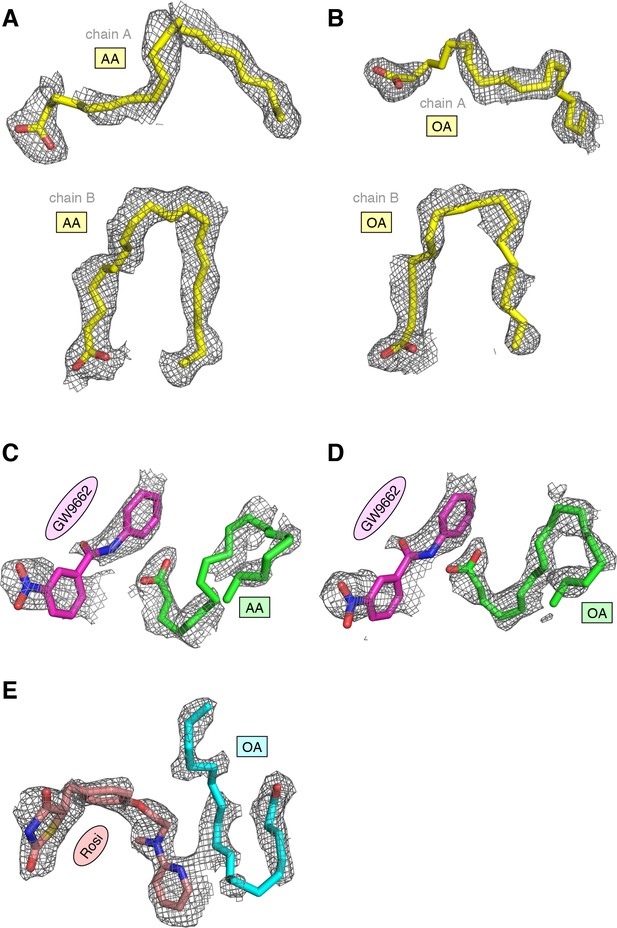
Omit map (2FO–FC, contoured at 1 σ) of the ligands in crystal structures of PPARγ LBD bound or cobound to (A) AA, (B) OA, (C) GW9662 and AA, (D) GW9662 and OA, and (E) rosiglitazone and OA.
https://doi.org/10.7554/eLife.43320.023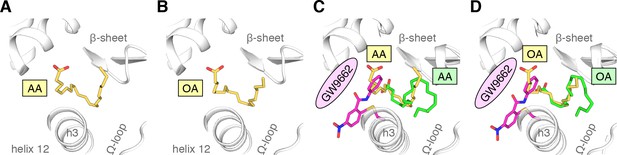
(A,B) UFA ligand binding poses in crystal structures of PPARγ LBD (chain B) bound to (A) AA and (B) OA.
(C,D) Structural overlays show that these chain B binding modes (yellow) overlaps with the binding GW9662-cobound fatty acid modes of (C) AA and (D) OA.
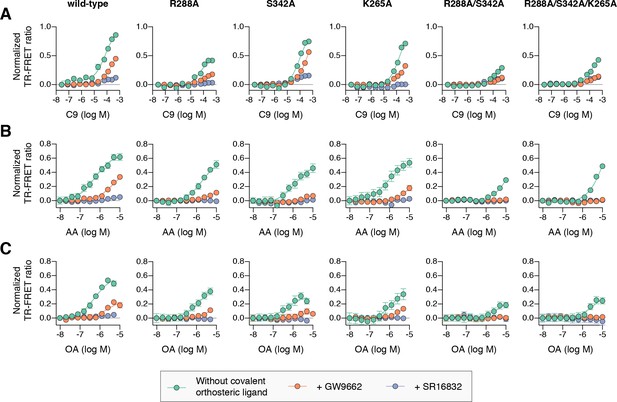
Effect of ligand cobinding of a covalent orthosteric synthetic ligand with MCFA or UFA ligands on PPARγ-coactivator recruitment.
TR-FRET biochemical assays showing concentration-dependent changes in the recruitment of a peptide derived from the TRAP220 coactivator for (A) C9, (B) AA, and (C) OA. Experiments were performed in the absence or presence of a covalent orthosteric synthetic ligand; and for wild-type PPARγ LBD or mutant variants; as indicated. Data plotted as the average and S.E.M. of three experimental replicates. Also see Figure 10—figure supplements 1–4.
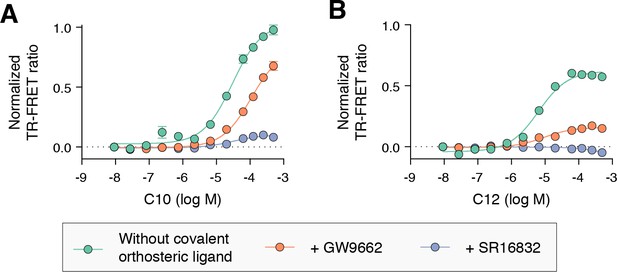
TR-FRET biochemical assays showing concentration-dependent changes in the recruitment of a peptide derived from the TRAP220 coactivator for (A) C10 and (B) C12.
Experiments were performed in the absence or presence of a covalent orthosteric synthetic ligand as indicated. Data plotted as the average and S.E.M. of three experimental replicates.
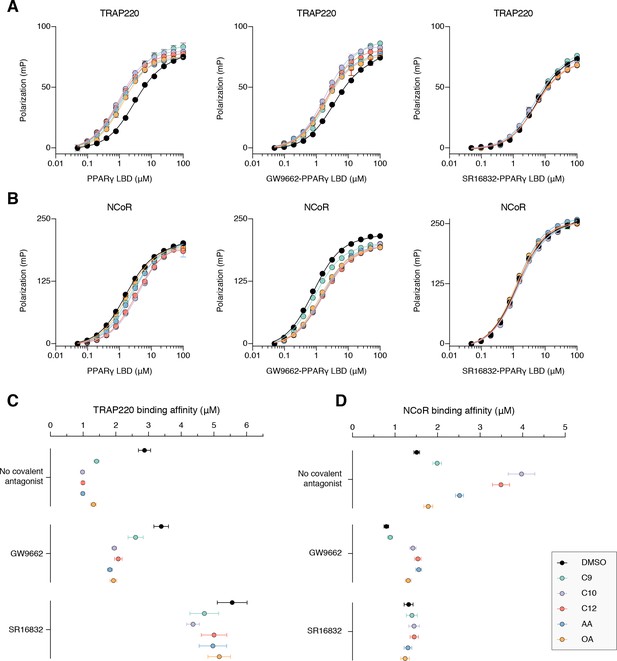
Fluorescence polarization (FP) assays performed using FITC-labeled peptides derived from (A) TRAP220 coactivator and (B) NCoR corepressor shows how coregulator affinities (C) and (D), respectively, are affected by cobinding of synthetic covalent ligands (GW9662 and SR16832) with MCFAs or UFAs.
Data in (A,B) plotted as the average and S.D. of three experimental replicates; data in (C,D) show the Kd values and fitted errors for the data in (A,B) that were fitted to a one site total binding equation.
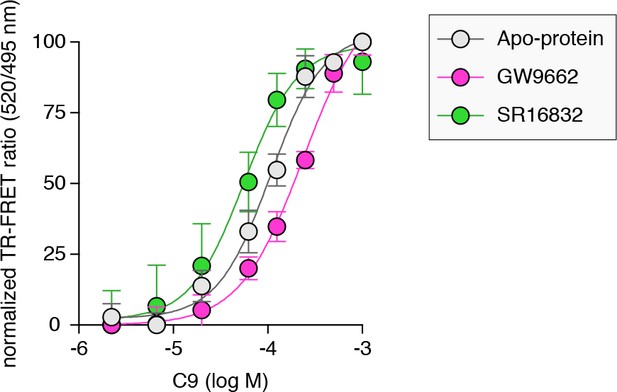
TRAP220-based TR-FRET assay performed in the absence or presence of covalent antagonists.
Data plotted as the average and S.D. of three experimental replicates.
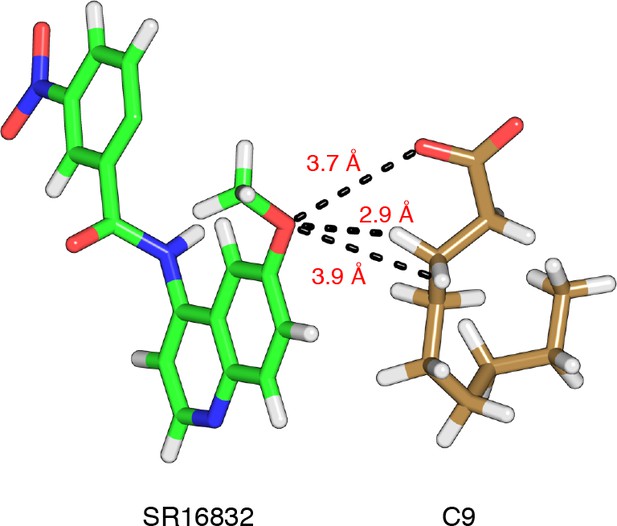
Hydrogen bonds/interactions between the SR16832 methoxy group and nonanoic acid.
https://doi.org/10.7554/eLife.43320.030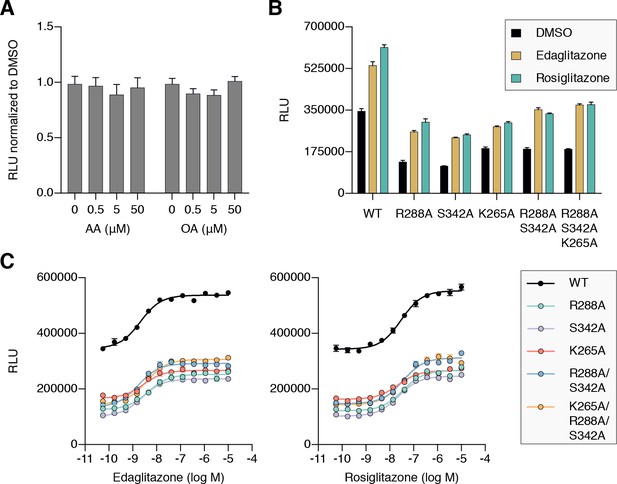
Cellular activation of PPARγ by TZDs may be influenced by cobinding of endogenous ligands.
(A) Luciferase reporter assay measuring full-length wild-type PPARγ transcription in HEK293T cells treated with exogenously added UFAs, which are endogenous ligands present in cells. (B,C) Luciferase reporter assay measuring full-length wild-type and mutant PPARγ transcription in HEK293T cells treated with ligand in (B) single-point (10 μM) and (C) dose-response format. Data plotted as the average and s.e.m. of four experimental replicates; dose response data were fit to a sigmoidal dose response equation. Fitted EC50 values are similar for wild-type and the mutant variants for edaglitazone (2–4 nM) and rosiglitazone (22–36 nM). Also see Figure 11—figure supplement 1 and 2.
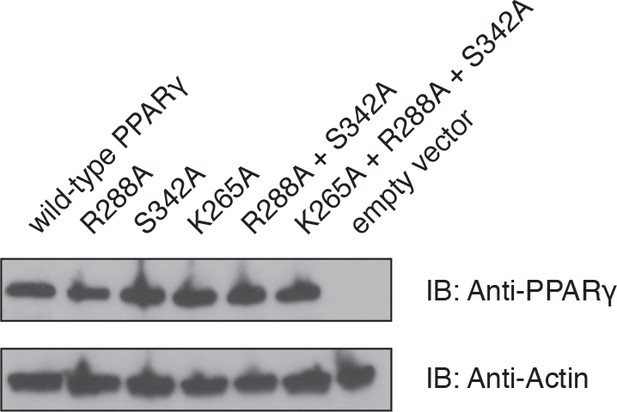
Western blot analysis protein levels in HEK293T cells transfected with wild-type or mutant full-length PPARγ expression plasmids.
https://doi.org/10.7554/eLife.43320.034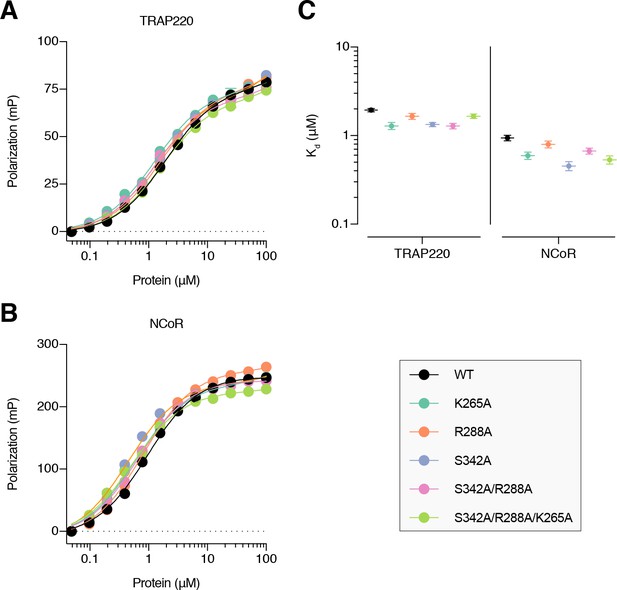
Fluorescence polarization (FP) assays performed using wild-type and mutant PPARγ LBD protein with FITC-labeled peptides derived from (A) TRAP220 coactivator and (B) NCoR corepressor shows (C) coregulator affinities are similar for the mutants.
Data in (A,B) plotted as the average and S.D. of three experimental replicates; data in (C) show the Kd values and fitted errors for the data in (A,B) that were fitted to a one site total binding equation.
Tables
X-ray crystallography data collection and refinement statistics.
https://doi.org/10.7554/eLife.43320.007| Edagtliazone (+ C9) | |
|---|---|
| Data collection | |
| Space group | I4 |
| Cell dimensions | |
| a, b, c (Å) | 128.74, 128.74, 93.67 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution | 33.36–2.1 (2.18–2.1) |
| Rpim | 0.049 (0.299) |
| I / σ(I) | 9.87 (2.83) |
| CC1/2 in highest shell | 0.798 |
| Completeness (%) | 99.31 (99.69) |
| Redundancy | 2.0 (2.0) |
| Refinement | |
| Resolution (Å) | 2.10 |
| No. of unique reflections | 44385 |
| Rwork/Rfree (%) | 18.2/21.6 |
| No. of atoms | |
| Protein | 4354 |
| Water | 473 |
| B-factors | |
| Protein | 27.30 |
| Ligand | 29.14 |
| Water | 33.73 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.18 |
| Ramachandran favored (%) | 96.28 |
| Ramachandran outliers (%) | 0.93 |
| PDB accession code | 5UGM |
-
*Values in parentheses indicate highest resolution shell.
X-ray crystallography data collection and refinement statistics.
https://doi.org/10.7554/eLife.43320.021| GW9662 (+ C9) | SR16832 (+ C9) | |
|---|---|---|
| Data collection | ||
| Space group | C 1 2 1 | C 1 2 1 |
| Cell dimensions | ||
| a, b, c (Å) | 92.57, 61.74, 118.38 | 92.61, 62.08, 118.45 |
| α, β, γ (°) | 90, 102.15, 90 | 90, 102.34, 90 |
| Resolution | 57.86–2.29 (2.37–2.29) | 45.24–2.73 (2.83–2.73) |
| Rpim | 0.029 (0.613) | 0.045 (0.338) |
| I / σ(I) | 11.90 (1.33) | 10.43 (2.00) |
| CC1/2 in highest shell | 0.757 | 0.832 |
| Completeness (%) | 98.94 (98.00) | 85.06 (74.34) |
| Redundancy | 6.6 (6.8) | 1.8 (1.8) |
| Refinement | ||
| Resolution (Å) | 2.29 | 2.73 |
| No. of unique reflections | 29660 | 15056 |
| Rwork/Rfree (%) | 24.9/31.4 | 19.9/28.1 |
| No. of atoms | ||
| Protein | 4145 | 4187 |
| Water | 243 | 60 |
| B-factors | ||
| Protein | 33.77 | 30.29 |
| Ligand | 45.19 | 39.84 |
| Water | 30.89 | 21.44 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (°) | 1.02 | 1.09 |
| Ramachandran favored (%) | 95.27 | 90.43 |
| Ramachandran outliers (%) | 1.58 | 1.95 |
| PDB accession code | 6AVI | 6AUG |
-
*Values in parentheses indicate highest resolution shell.
X-ray crystallography data collection and refinement statistics.
https://doi.org/10.7554/eLife.43320.025| Arachidonic acid | Oleic acid | GW9662 + Arachidonic acid | GW9662 + Oleic acid | Rosiglitazone + Oleic acid | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C 1 2 1 | C 1 2 1 | C 1 2 1 | C 1 2 1 | C 1 2 1 |
| Cell dimensions | |||||
| a, b, c (Å) | 93.04, 62.16, 118.96 | 92.93, 62.17, 119.32 | 92.88, 62.10, 119.19 | 92.78, 61.66, 118.63 | 92.83, 61.83, 118.72 |
| α, β, γ (°) | 90, 102.38, 90 | 90, 102.20, 90 | 90, 101.90, 90 | 90, 102.15, 90 | 90, 102.34, 90 |
| Resolution | 44.97–2.10 (2.12–2.10) | 38.88–1.95 (2.02–1.95) | 38.87–2.2 (2.279–2.2) | 39.51–2.2 (2.279–2.2) | 57.99–2.24 (2.32–2.24) |
| Rpim | 0.039 (0.429) | 0.036 (0.471) | 0.016 (0.277) | 0.014 (0.283) | 0.045 (0.469) |
| I / σ(I) | 10.06 (1.65) | 8.16 (1.34) | 17.06 (2.52) | 17.74 (2.57) | 10.32 (3.02) |
| CC1/2 in highest shell | 0.766 | 0.785 | 0.892 | 0.976 | 0.761 |
| Completeness (%) | 98.42 (95.47) | 95.30 (93.96) | 98.34 (97.39) | 98.24 (97.93) | 97.98 (86.12) |
| Redundancy | 1.9 (1.9) | 1.7 (1.6) | 2.0 (2.0) | 2.0 (2.0) | 3.2 (3.1) |
| Refinement | |||||
| Resolution (Å) | 2.10 | 1.95 | 2.20 | 2.20 | 2.24 |
| No. of unique reflections | 38363 | 46478 | 33474 | 32923 | 31810 |
| Rwork/Rfree (%) | 21.3/25.7 | 22.5/27.3 | 22.6/26.2 | 22.5/26.6 | 24.4/28.5 |
| No. of atoms | |||||
| Protein | 4102 | 4102 | 4102 | 4118 | 4081 |
| Water | 411 | 502 | 244 | 232 | 224 |
| B-factors | |||||
| Protein | 27.08 | 24.94 | 32.75 | 32.18 | 34.00 |
| Ligand | 43.30 | 40.12 | 46.47 | 50.57 | 48.95 |
| Water | 32.32 | 30.69 | 34.42 | 31.61 | 35.11 |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.008 | 0.009 | 0.009 | 0.010 | 0.008 |
| Bond angles (°) | 1.15 | 1.20 | 1.19 | 1.34 | 0.97 |
| Ramachandran favored (%) | 98.19 | 99.00 | 97.59 | 94.82 | 97.18 |
| Ramachandran outliers (%) | 0.60 | 0.20 | 0.40 | 1.99 | 1.01 |
| PDB accession code | 6MCZ | 6MD0 | 6MD2 | 6MD1 | 6MD4 |
-
*Values in parentheses indicate highest resolution shell.
ITC analysis of TRAP220 peptide titrated into PPARγ LBD.
https://doi.org/10.7554/eLife.43320.031| Ligand | log Ka | Kd (μM) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | TΔs (kcal mol−1) | n (from two replicates) |
|---|---|---|---|---|---|---|
| DMSO | 4.60 (4.53, 4.66) | 25.29 (21.65, 29.77) | −6.271 | −10.53 (−11.58,–9.70) | −4.257 | 0.907, 1.088 |
| C9 (3x) | 5.71 (5.61, 5.82) | 1.94 (1.53, 2.46) | −7.793 | −10.40 (−11.10,–9.81) | −2.612 | 1.059, 1.087 |
| OA (2x) | 5.90 (5.82, 5.97) | 1.27 (1.07, 1.51) | −8.043 | −9.24 (−10.18,–8.31) | −1.192 | 1.276, 1.229 |
| Edaglitazone | 5.90 (5.84, 5.96) | 1.26 (1.10, 1.42) | −8.051 | −10.04 (−10.24,–9.84) | −1.992 | 0.907, 0.863 |
| Rosiglitazone | 5.89 (5.88, 5.94) | 1.28 (1.13, 1.43) | −8.042 | −8.88 (−9.04,–8.72) | −0.838 | 0.907, 0.909 |
| Edaglitazone + C9 | 6.20 (6.10, 6.30) | 0.63 (0.50, 0.78) | −8.457 | −9.71 (−9.98,–9.44) | −1.254 | 0.840, 0.922 |
| Rosiglitazone + C9 | 6.24 (6.16, 6.34) | 0.57 (0.46, 0.69) | −8.523 | −9.12 (−9.38,–8.87) | −0.596 | 0.939, 0.952 |
| Rosiglitazone + OA | 6.16 (6.02, 6.31) | 0.69 (0.49, 0.95) | −8.406 | −7.92 (−8.45,–7.46) | 0.487 | 1.210, 1.258 |
-
Data represent values from an unbiased global fitting of two independent ITC experiments per condition. The 68.3% confidence interval from global fitting listed as italicized values in parentheses when applicable. Stoichiometry (n value) is listed for each independent experiment. Ligands were present at the following molar equivalents: one equiv. (edaglitazone and rosiglitazone), two equiv. (OA), or three equiv. (C9).
-
Table 4—source data 1
Thermograms and normalized plotted data from ITC titration of TRAP220 peptide into PPARγ LBD.
Two replicate measurements (green and pink data) per ligand-bound condition (molar equivalents of 1X for rosiglitazone or edaglitazone, 2X for OA, and 3X for C9) were used for the unbiased global ITC analysis.
- https://doi.org/10.7554/eLife.43320.032
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43320.036





