Guy1, a Y-linked embryonic signal, regulates dosage compensation in Anopheles stephensi by increasing X gene expression
Figures
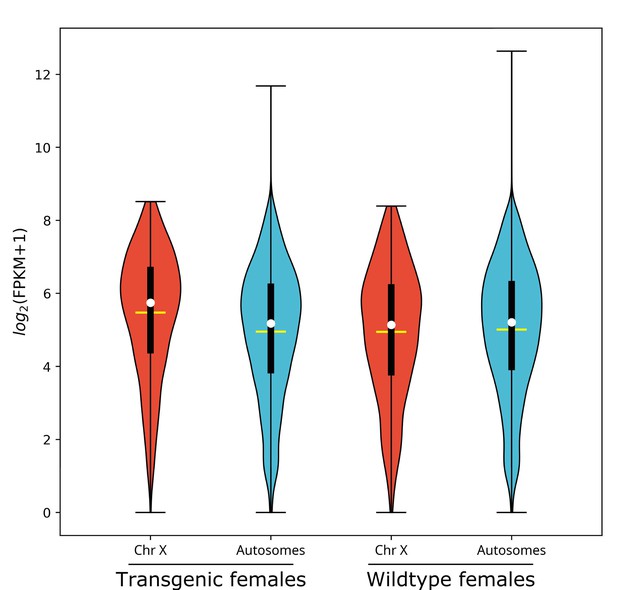
X chromosome median gene expression is up-regulated in Guy1 transgenic females of line nGuy1_2.
Violin plot shows gene expression as log2(FPKM +1) from the X chromosome and autosomes from transgenic and wild-type females. Transcripts under a relatively low expression level (FPKM <1) are excluded. The width of the violin plots shows the density of transcripts at different expression values. The white dot denotes the median of each group, while the yellow horizontal bar denotes the mean. The bottom and top of the vertical thick black bars represent the first and third quartiles, respectively. In transgenic females the median expression level of the X chromosome is significantly higher than the autosomes according to the two-tailed Wilcoxon tests, while no significant difference is seen in wild-type females. Median expression value comparisons between the X chromosome and autosomes for transgenic and wild-type females are 53.6 vs. 36.6 (p=2.2e−16), 35.4 vs. 36.9 (p=0.31), respectively. One of the four transgenic female replicates (TF1) was excluded from the above analysis on the basis of PCA (Figure 1—figure supplement 1). TF1 had a lower overall gene expression than the other replicates (Supplementary file 3). However, the median gene expression of X-linked genes is still significantly higher than that of the autosomes in TF1 (Table 3). An independent experiment validated these results in females, and for additional male samples included, there was no significant difference in the median gene expression between the X chromosome and autosomes in transgenic males or non-transgenic males (Figure 1—figure supplement 2). All median FPKM expression value comparisons with statistical significance can be found in Supplementary file 2.
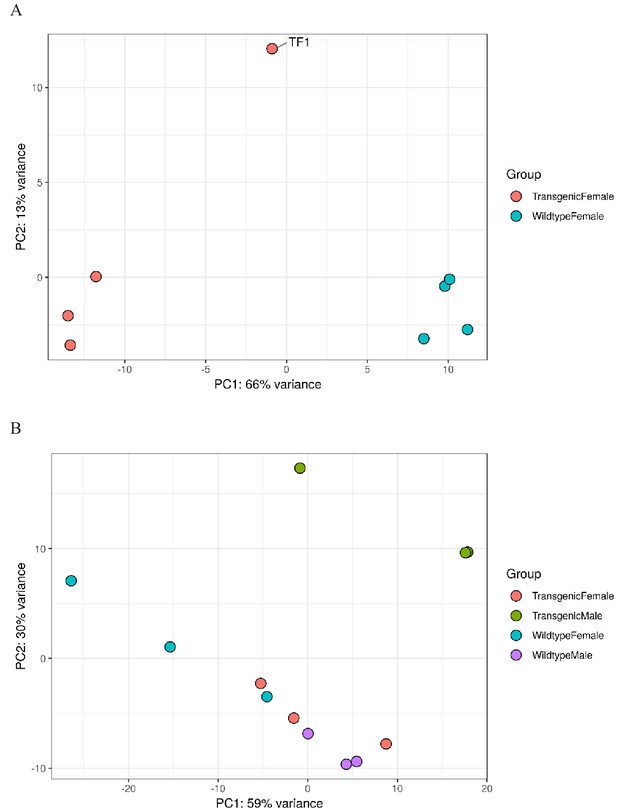
Principal component analysis of replicates from Experiment A (Panel A, two genotypes from nGuy1_2) and Experiment B (Panel B, four genotypes from nGuy1_1).
https://doi.org/10.7554/eLife.43570.005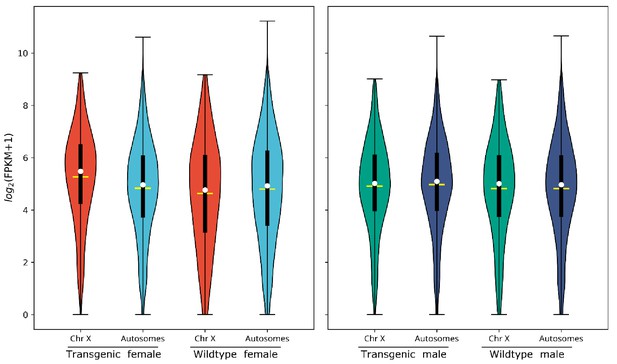
X chromosome median gene expression is up-regulated in Guy1 transgenic females from line nGuy1_1.
https://doi.org/10.7554/eLife.43570.006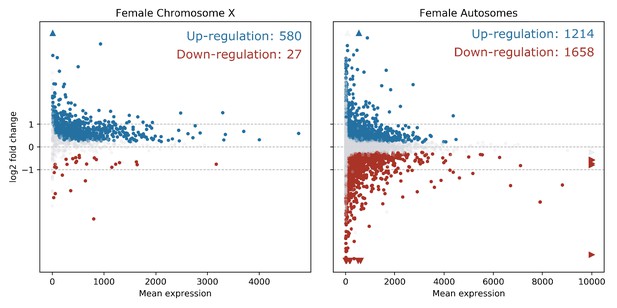
X chromosome genes are upregulated in Guy1 transgenic females of line nGuy1_2.
MA Plots show differentially expressed genes on the X chromosome and the autosomes in Guy1 transgenic female compared to wild-type female siblings. Each dot represents one gene. Blue dots denote upregulated genes, the red dots denote downregulated genes, and grey dots are not differentially expressed (BH-adjusted p>0.1). Colored arrows indicate values beyond the range of the plot. Genes are significantly upregulated on the X chromosome in transgenic females compared to the autosomes, which is supported by a Chi-square test (p=2.20e−16, Supplementary file 5). A repeat experiment with essentially the same results that also includes male samples is shown in Figure 2—figure supplement 1.
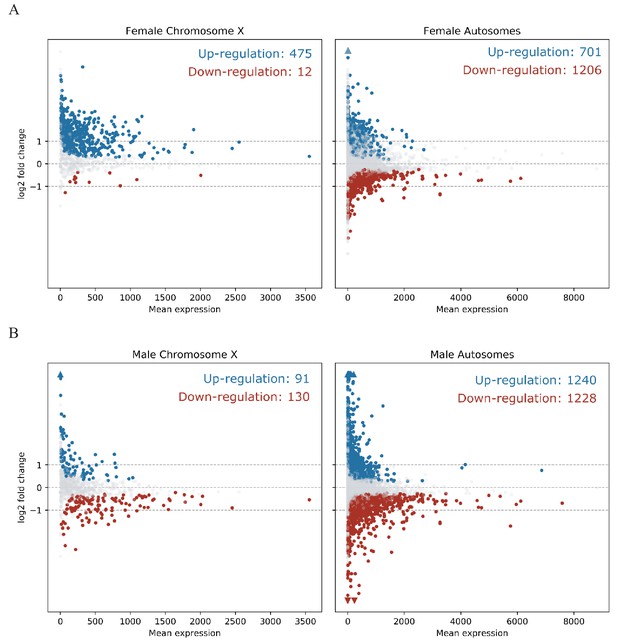
X-linked genes are upregulated in Guy1 transgenic females from line nGuy1_1.
https://doi.org/10.7554/eLife.43570.009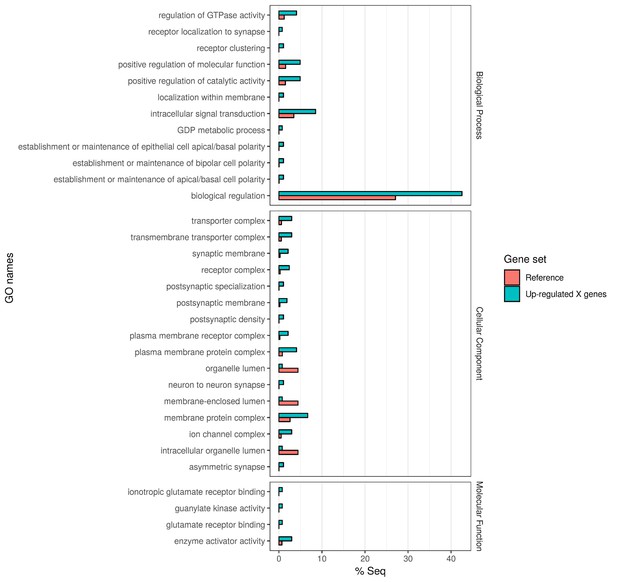
Gene Ontology enrichment of the 382 X chromosome genes that are upregulated in the Guy1-transgenic females from both nGuy1_1 and nGuy1_2 lines.
There are 475 and 580 upregulated X-linked genes identified by DESeq2 in the transgenic females from line nGuy1_1 and nGuy1_2, respectively (Supplementary file 4 and Supplementary file 5). Among these, 382 genes were found in common (Supplementary file 4). These 382 genes were mapped to GO names and a two-tailed Fisher’s exact test was performed to detect enrichment of GO names against AsteI2.2 transcripts under FDR of 0.05 using Blast2GO (V5.1.1) (Götz et al., 2008).
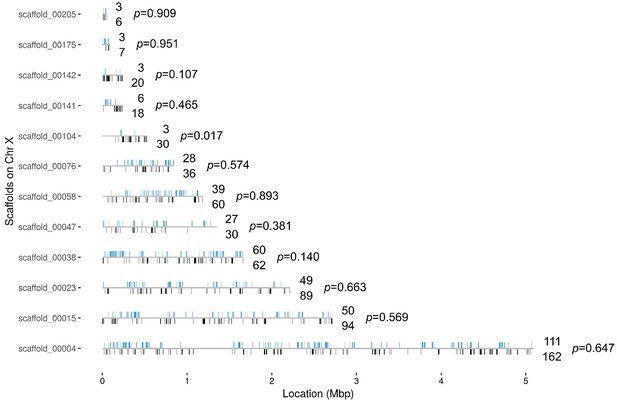
Distribution of the 382 X chromosome genes that are upregulated in the Guy1-transgenic females from both nGuy1_1 and nGuy1_2 lines.
There is not yet a chromosomal level assembly for Anopheles stephensi. The location (from start to end) of the 996 X genes are mapped on the 12 X-scaffolds (Supplementary file 4). Blue bars above the line indicates the position of genes that are upregulated and the number above the line indicates the number of upregulated genes in each scaffold. Black bars underneath the line indicates the position of other genes and the number below the line indicates the number of other genes in each scaffold. There are only seven X genes that are down-regulated in the Guy1-transgenic females from both lines (Supplementary file 4 sheet 4). The p-values show the results of a Chi-square test of each scaffold for deviation from the 382/996 ratio. We also broke scaffolds 00023, 00015, and 00004 into 2, 3, and 5 evenly divided segments, respectively, to perform the same Chi-square test. Segment 2 of scaffold_00004 showed a p-value of 0.09 while all other nine segments showed p-values between 0.34 and 0.99. Thus, although minor local biases exist, the 382 upregulated genes are widely distributed across the X chromosome.
Tables
Adult screening of Guy1-expressing lines nGuy1_2 and bGuy1tC demonstrate 100% female lethality.
https://doi.org/10.7554/eLife.43570.002| Line | TM (%) | TF | WM (%) | WF (%) | Total progeny | |
|---|---|---|---|---|---|---|
| nGuy1_2 | 1970 (35.5) | 0 | 1800 (32.5) | 1776 (32.0) | 5546 | |
| bGuy1tC | 1506 (37.0) | 0 | 1340 (32.9) | 1223 (30.1) | 4069 | |
| 3476 | 0 | 3140 | 2999 | 9615 | ||
-
TM stands for transgenic male and WM stands for wild type male. TF stands for transgenic female and WF stands for wild type female.
For these screening results line nGuy1_2 was at generation 59, 60 and line bGuy1tC was at generation 16, 17.
Monitoring 1st instar larvae indicates delayed transgenic female hatching and female lethality.
https://doi.org/10.7554/eLife.43570.003| Transgenic line | Genotype | 42 hr PO | 50 hr PO | 58 hr PO | 66 hr PO | 70 hr PO | 82 hr PO | Adults (%) |
|---|---|---|---|---|---|---|---|---|
| nGuy1_2 | TF | 0 | 0 | 22 | 20 | 2 | 0 | 0 |
| WF | 169 | 199 | 201 | N/A | N/A | N/A | 168 (32.0) | |
| TM | 181 | 206 | 209 | N/A | N/A | N/A | 175 (33.3) | |
| WM | 189 | 219 | 222 | N/A | N/A | N/A | 182 (34.7) | |
| bGuy1tC | TF | 0 | 0 | 16 | 16 | 12 | 0 | 0 |
| WF | 159 | 166 | 166 | N/A | N/A | N/A | 122 (33.2) | |
| TM | 162 | 169 | 170 | N/A | N/A | N/A | 136 (37.0) | |
| WM | 139 | 154 | 154 | N/A | N/A | N/A | 110 (29.9) |
-
TF stands for transgenic female and WF stands for wild type female. TM stands for transgenic male and WM stands for wild-type male. PO stands for Post Oviposition.
Numbers from 42 h – 82 h PO represent larval counts.
-
N/A - after 58 hr all larvae had hatched and only transgenic female larvae were monitored for death.
Similar experiments have been repeated more than three times for each line with essentially the same results.
The median level gene expression of genes on X chromosome and autosomes of individual replicates of line nGuy1_2.
https://doi.org/10.7554/eLife.43570.007| Sample* | Chromosome X | Autosomes | P value† |
|---|---|---|---|
| TF1 | 38.12126 | 33.37074 | 0.0012021 |
| TF2 | 54.33247 | 36.23512 | 0.0000000 |
| TF3 | 53.00118 | 36.08971 | 0.0000000 |
| TF4 | 54.14014 | 34.74229 | 0.0000000 |
| WF1 | 38.07142 | 38.49541 | 0.7098231 |
| WF2 | 36.15963 | 36.88966 | 0.6335401 |
| WF3 | 33.74510 | 35.75736 | 0.2684612 |
| WF4 | 30.98920 | 32.89602 | 0.0860183 |
-
*TF stands for transgenic female and WF stands for wild type female. Transcripts under a relatively low expression level (FPKM <1) are excluded. Results of different FPKM cutoffs are provided in Supplemental File S3.
†The p values were calculated based on the two-tailed two-sample Wilcoxon rank sum test.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Anopheles stephensi) | Guy1 | PMID: 23683123; PMID: 27644420 | JX174417; AFS60403 | |
| Genetic reagent (Anopheles stephensi) | X-linked CFP (cyan fluorescent protein) line | PMID: 20113372 | ||
| Biological sample (Anopheles stephensi, Indian) | The transcriptome of Guy1-transgenic Anopheles stephensi L1 instar (TaxID: 30069) | this paper | PRJNA503140 (NCBI Sequence Read Archive) | |
| Recombinant DNA reagent | bGuy1tC; nGuy1 | this paper; PMID: 27644420 | ||
| Recombinant DNA reagent | piggyBac transformation vector and helper | PMID: 11151299 | ||
| Sequence- based reagent | An. stephensi reference genome (Version AsteI2); An. stephensi transcripts (Version AsteI2.2) | PMID: 25244985; can also be accessed at Vectorbase.org | ||
| Sequence- based reagent | Strep-tag II | PMID: 17571060 | ||
| Commercial assay or kit | Quick-RNA Miniprep kit | Zymo Research | R1054 | |
| Software, algorithm | HISAT (V2.1.0) | PMID: 25751142 | ||
| Software, algorithm | SAMtools | PMID: 19505943 | ||
| Software, algorithm | MarkDuplicates | https://broadinstitute.github.io/picard/ | ||
| Software, algorithm | StringTie (V1.3.3) | PMID: 25690850 | ||
| Software, algorithm | Python script | this paper; https://gist.github.com/yangwu91/ | 79b74035465978e 78d41af4236597ff1 | |
| Software, algorithm | Matplotlib Python Package | DOI:10.1109 /Mcse.2007.55 | ||
| Software, algorithm | GenomicAlignments R package | PMID: 23950696 | ||
| Software, algorithm | DESeq2 | PMID: 25516281 |
Additional files
-
Supplementary file 1
Spearman correlation analyses of the two RNA-Seq experiments.
- https://doi.org/10.7554/eLife.43570.012
-
Supplementary file 2
Analyses of median FPKMs of X-linked genes vs. autosomal genes.
This file shows results from analyses of pooled biological replicates.
- https://doi.org/10.7554/eLife.43570.013
-
Supplementary file 3
Analyses of median FPKMs of X-linked genes vs. autosomal genes of Guy1 transgenic and sibling wild type males and females: Individual replicates.
- https://doi.org/10.7554/eLife.43570.014
-
Supplementary file 4
DESeq2 output, overlap of differentially expressed genes, scaffold positions of up-regulated and other X-linked genes (This file includes five sheets of an Excel workbook, uploaded separately)
- https://doi.org/10.7554/eLife.43570.015
-
Supplementary file 5
Significantly upregulated and downregulated genes on chromosome X and autosomes between Guy1 transgenic and wild-type sibling males and females.
- https://doi.org/10.7554/eLife.43570.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43570.017





