Requirements for RNA polymerase II preinitiation complex formation in vivo
Figures
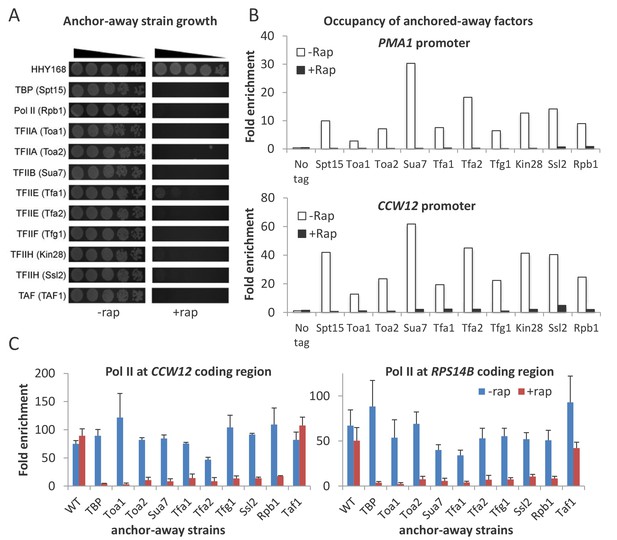
Conditional depletion of GTFs causes severe growth and transcriptional defects.
(A) Growth of the indicated anchor-away cells (5-fold serial dilutions) in the presence or absence of rapamycin. (B) Occupancy of the indicated FRB-tagged GTFs at the PMA1 and CCW12 promoters in the corresponding strains and an untagged control strain in the presence of absence of rapamycin. (C) Pol II occupancy at the CCW12 and RPS14B coding regions in the indicated strains grown in the presence or absence of rapamycin. Error bars represent the standard error of at least three independent experiments.
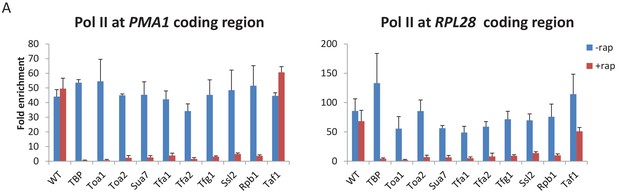
Conditional depletion of GTFs causes severe growth and transcriptional defects.
Pol II occupancy at the PMA1 and RPL28 coding regions in the indicated strains grown in the presence or absence of rapamycin.
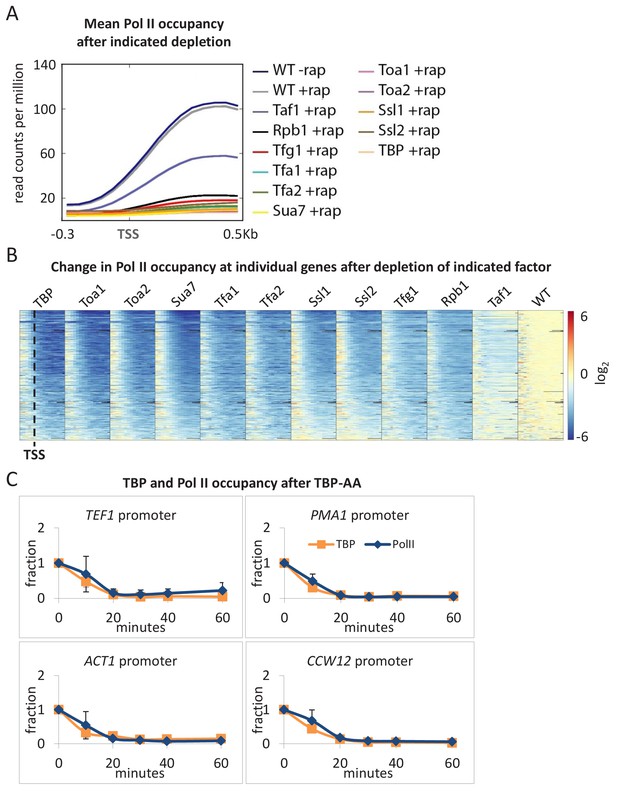
All GTFs are generally required for ongoing Pol II transcription.
(A) Mean Pol II occupancy averaged over 453 well-transcribed genes (metagene analysis) in strains depleted (+rap) for the indicated factor and in the parental (WT) strain (±rap). Partial reduction is observed only for the TAF1-depleted strain. (B) Pol II occupancy at individual genes (the same set of 453 genes ordered from top to bottom by expression level in WT) in strains depleted for the indicated factor. For each gene, the log2 change in Pol II occupancy after depletion is indicated according to the red/blue scale. (C) TBP and Pol II occupancies at the indicated promoters in the TBP-depletion strain at various times after rapamycin treatment. Error bars represent the standard error of at least three independent experiments.
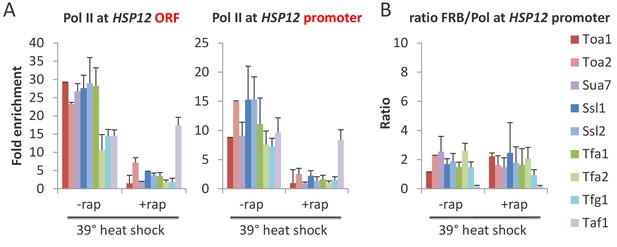
All GTFs are required for transcriptional induction upon heat shock.
(A) Mean Pol II occupancy at the HSP12 coding region (ORF) and promoter in strains depleted (or not) for the indicated factor and then induced for 15 min by shifting to 39°C. (B) FRB-tagged GTF:Pol II occupancy ratio at the induced HSP12 promoter in cells pretreated or not with rapamycin to deplete the indicated factors.
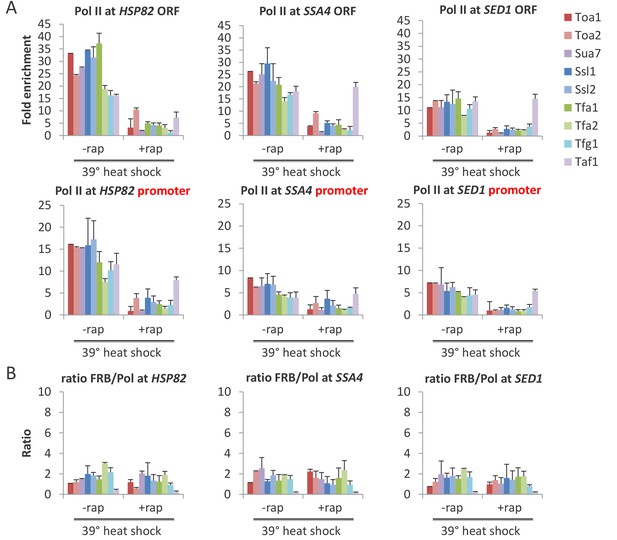
All GTFs are required for transcriptional induction upon heat shock.
(A) Pol II occupancy at the coding region (ORF) and promoter of the indicated genes in strains depleted (or not) for the indicated factor and then induced for 15 min by shifting to 39°C. (B) FRB-tagged GTF:Pol II occupancy ratio at the induced promoter in cells pretreated or not with rapamycin to deplete the indicated factors.
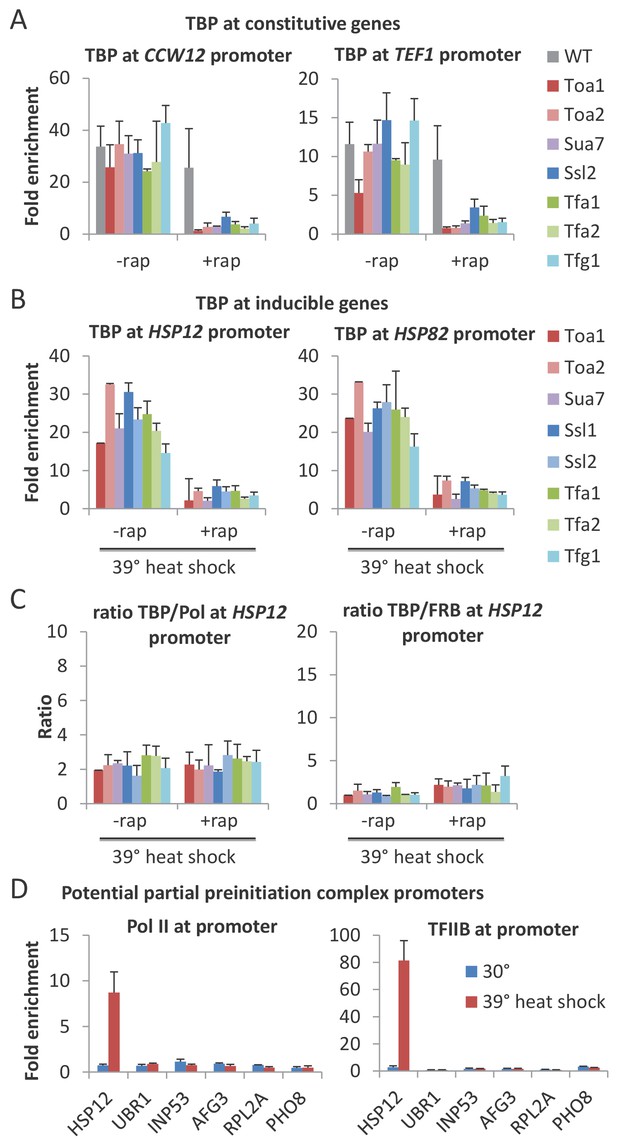
All GTFs are required for TBP occupancy, and hence PIC stability/formation.
(A) TBP occupancy at the CCW12 and TEF1 promoters in strains depleted (+rap) or not (-rap) for the indicated factor. (B) TBP occupancy at the HSP12 and HSP82 promoters in strains depleted (+rap) or not (-rap) for the indicated factor and subject to a heat shock. (C) TBP:Pol II and TBP:FRB-tagged GTF occupancy ratios at the induced HSP12 promoter in cells pretreated or not with rapamycin to deplete the indicated factors. (D) Pol II and TFIIB occupancies at the indicated promoters previously reported to have partial PICs (Zanton and Pugh, 2006) under normal (blue) and heat shock induction (red).
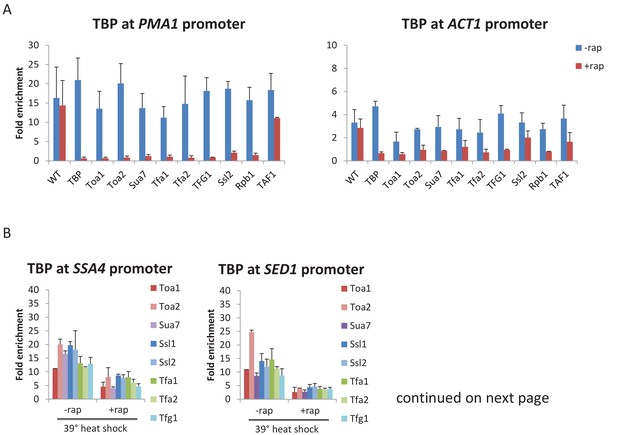
All GTFs are required for TBP occupancy, and hence PIC stability/formation.
(A) TBP occupancy at the PMA1 and ACT1 promoters in strains depleted (+rap) or not (-rap) for the indicated factor. (B) TBP occupancy at the SSA4 and SED1 promoters in strains depleted (+rap) or not (-rap) for the indicated factor and subject to a heat shock. (C) TBP:Pol II and TBP:FRB-tagged GTF occupancy ratios at the induced promoters in cells pretreated or not with rapamycin to deplete the indicated factors.
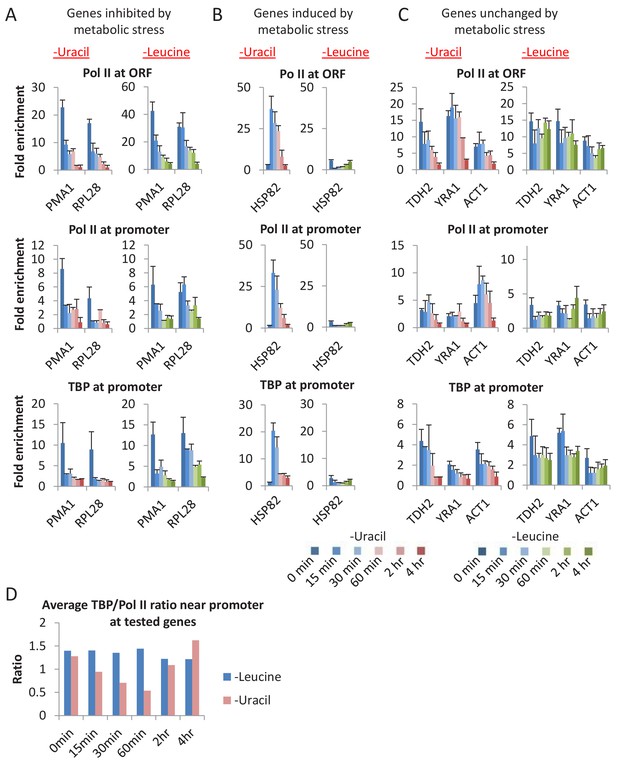
A general loss of PICs in cells depleted for uracil.
(A) Pol II (ORF and promoter) and TBP occupancies at promoters (PMA1 and RPL28) of genes inhibited by metabolic stress in cells depleted for uracil or leucine for various times (color scale). (B) Similar analysis for HSP82, a gene induced by various metabolic stresses. (C) Similar analysis for genes TDH2, YRA1, and ACT1, genes unchanged upon metabolic stress. (D) Average TBP/Pol II ratio at all tested promoters at various times after depletion of leucine (blue) or uracil (red). See Figure 5—figure supplement 1D for individual tested promoters.
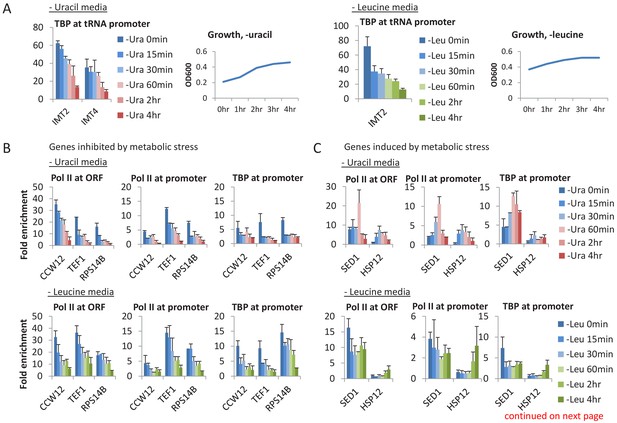
A general loss of PICs in cells depleted for uracil.
(A) TBP occupancy at Pol III (tRNA) promoters, as well as growth curves for cultures depleted of uracil or leucine. (B) Pol II (ORF and promoter) and TBP occupancies at promoters of genes inhibited by metabolic stress in cells depleted for uracil or leucine for various times (color scale). (C) Similar analysis for SED1 and HSP12, which are induced by various metabolic stresses. (D) TBP/Pol II ratio at nine tested promoters at various times after depletion of leucine or uracil.
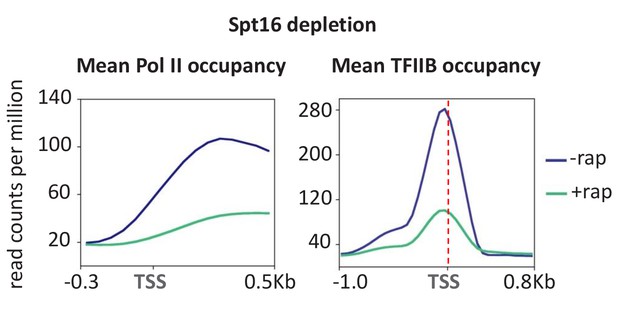
Depletion of Spt16 subunit of FACT reduces transcription and PIC formation.
Mean Pol II occupancy and TFIIB occupancy averaged over 453 well-transcribed genes and promoters before and after Spt16 depletion.
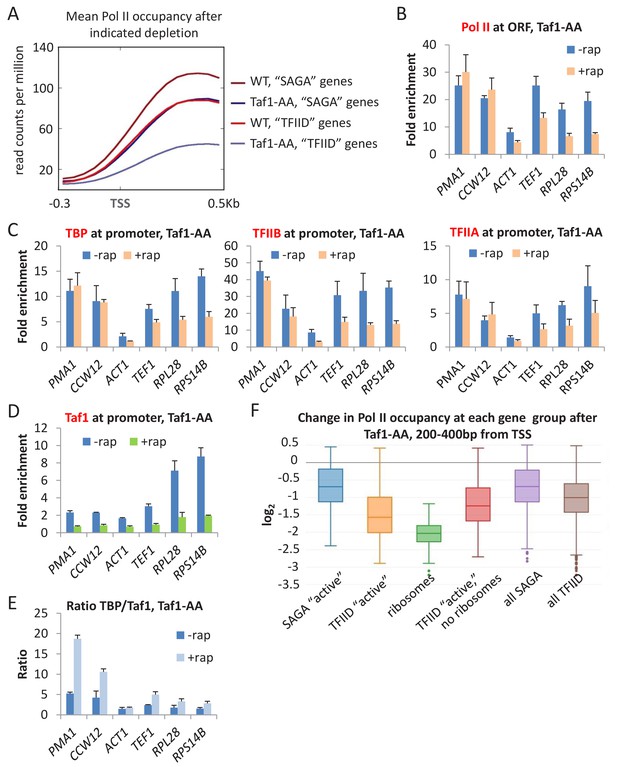
Taf1 depletion selectively affects TFIID-dependent genes.
(A) Mean Pol II occupancy averaged over 453 well-transcribed ‘SAGA’ or ‘TFIID’ genes in parental or Taf1-depleted strains. (B) Pol II (C) TBP, TFIIB, and TFIIA, and (D) Taf1 occupancies at ‘SAGA’ (PMA1 and CCW12) or ‘TFIID’ (ACT1, TEF1, RPL28, RPS14B) coding regions in the Taf1-depletion strain treated or untreated with rapamycin. (E) TBP:TAF1 occupancy ratios in the Taf1-depletion strain treated or untreated with rapamycin. (F) Log2 change in Pol II occupancy (measured at +200–400 from the TSS) for the indicated gene classes upon Taf1 depletion. ‘Active’ genes are the top 10% of transcribed genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories. Ribosomes include only the ribosomal genes from the TFIID-dependent category. ‘TFIID active, no ribosomes’ are the remainder of the top 10% transcribed genes in the TFIID-dependent category after ribosomal genes have been removed. ‘All’ genes are the entire set of ~5000 s. cerevisiae genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories.
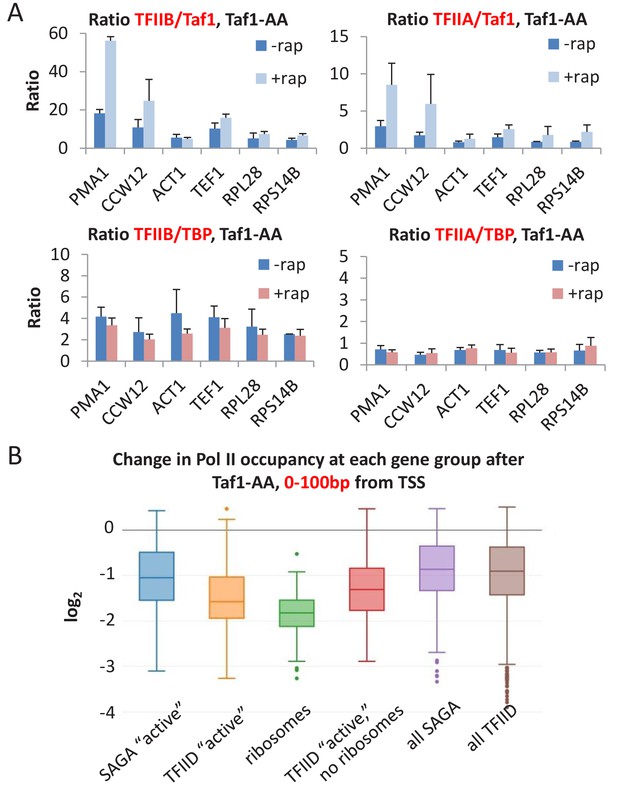
Taf1 depletion selectively affects TFIID-dependent genes.
(A) TFIIB:Taf1, TFIIA:Taf1, TFIIB:TBP, and TFIIA:TBP occupancy ratios in the Taf1-depletion strain treated or untreated with rapamycin. (B) Log2 change in Pol II occupancy (measured at 0 to +100 bp from the TSS) for the indicated gene classes upon Taf1 depletion. ‘Active’ genes are the top 10% of transcribed genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories. Ribosomes include only the ribosomal genes from the TFIID-dependent category. ‘TFIID active, no ribosomes’ are the remainder of the top 10% transcribed genes in the TFIID-dependent category after ribosomal genes have been removed. ‘All’ genes are the entire set of ~5000 s. cerevisiae genes, broken up into ‘TATA’ and ‘TFIID’-dependent categories.
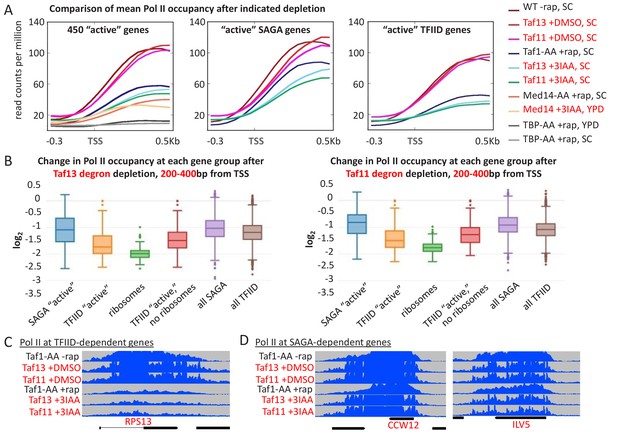
Analysis of Taf11 and Taf13 depletion in SC medium using published data (Warfield et al., 2017) shows selective effects at TFIID-dependent genes.
(A) Mean Pol II occupancy averaged over 453 well-transcribed ‘SAGA’ or ‘TFIID’ genes in parental or Taf1-depleted anchor-away strains (black; present study) as well as Taf11- and Taf13-depleted auxin-degron strains (red) treated with DMSO or with 3IAA (Warfield et al., 2017). (B) Log2 change in Pol II occupancy (measured at +200–400 from the TSS) for the indicated gene classes upon Taf13 and Taf11 depletion. ‘Active’ genes are the top 10% of transcribed genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories. Ribosomes include only the ribosomal genes from the TFIID-dependent category. ‘TFIID active, no ribosomes’ are the remainder of the top 10% transcribed genes in the TFIID-dependent category after ribosomal genes have been removed. ‘All’ genes are the entire set of ~5000 s. cerevisiae genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories. (C and D) Screenshots in the Integrated Genome Browser of Pol II levels at the indicated TFIID-dependent and SAGA-dependent genes before and after anchor-away depletion of Taf1 or before and after auxin-degron depletion of Taf11 and Taf13.
Additional files
-
Supplementary file 1
List of strains.
- https://doi.org/10.7554/eLife.43654.015
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43654.016





