Structure and mechanism of a Hypr GGDEF enzyme that activates cGAMP signaling to control extracellular metal respiration
Figures
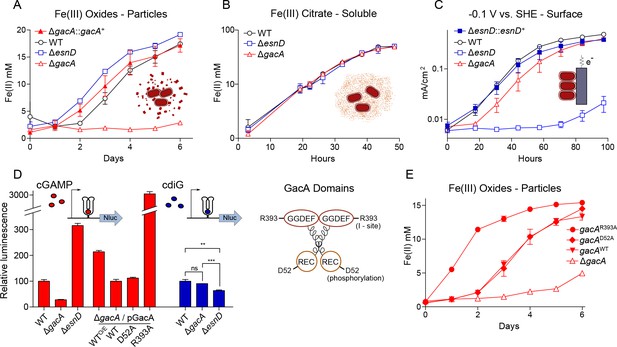
GacA synthesizes cyclic GMP-AMP and controls Fe(III) particle respiration in vivo, whereas EsnD synthesizes cyclic di-GMP and controls electrode respiration.
(A) G. sulfurreducens ΔgacA deletion strain is defective in the reduction of insoluble Fe(III) oxide particles (open triangles). Re-expressing gacA in the Tn7 site on the chromosome (filled triangles) restores Fe(III) reduction. An ΔesnD deletion strain (squares) exhibits no lag in Fe(III) oxide reduction compared to 2 d after inoculation for WT (circles). (B) Soluble Fe(III) citrate reduction is unaffected in either ΔgacA (triangles) and ΔesnD (squares) strains relative to WT (circles). (C) EsnD is required for robust electrode reduction (open squares) and re-expressing esnD in the Tn7 site on the chromosome (filled squares) restores electrode respiration to WT (circles) levels. GacA is not required for electrode respiration (triangles). (D) Strains containing riboswitches driving Nanoluc luciferase, reporting cellular levels of cGAMP (red bars) and cdiG (blue bars). The ΔgacA strains express GacA variants from the Tn7 chromosomal site (also see cartoon). Deleting gacA reduces cGAMP-dependent reporter levels by ~80% but has no effect on cdiG reporter levels. Deleting esnD reduces cdiG levels by ~20% and increases cGAMP levels to 3x WT. The Rec domain variant D52A produces cGAMP at levels comparable to WT, while the I-site variant R393A increases cGAMP levels to 30x WT. (E) Reduction of Fe(III) oxide particles in the ΔgacA deletion strain is rescued by re-expressing WT GacA and D52A variants, and exhibits significantly increased rate for the R393A variant that overproduces cGAMP. Representative biological replicates are shown for Fe(III) oxide reduction (n = 3), Fe(III) citrate reduction (n = 3), and electrode growth (n = 4). Nanoluc assays were performed in biological replicates (n = 3) in panel D. P values in panel D: ns > 0.05; **=0.001–0.01; ***<0.001. All error bars represent standard deviations.
-
Figure 1—source data 1
Growth rates and nanoluc reporter data.
- https://doi.org/10.7554/eLife.43959.004
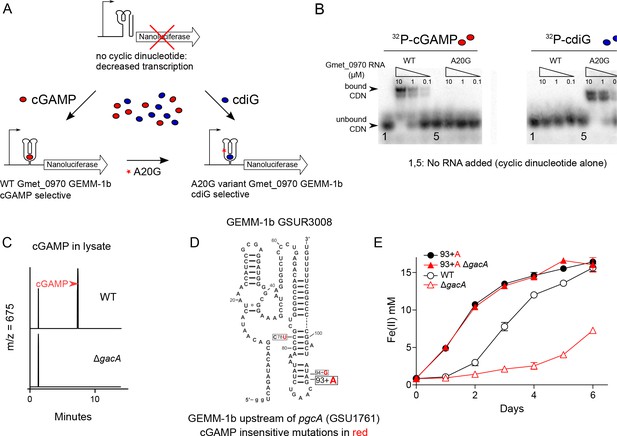
Nanoluciferase fusion to WT cGAMP riboswitch and engineered cdiG riboswitch for in vivo cyclic dinucleotide analysis; riboswitch GSUR3008 variants that control pgcA (GSU1761) expression in Geobacter sulfurreducens.
(A) Expression of nanoluciferase is induced by binding of cyclic dinucleotides to GEMM-1b riboswitch. An A to G variant in nucleotide 20 in the Gmet_0970 GEMM-1b riboswitch changes cGAMP selectivity to cdiG. (B) The WT and A20G variant of Gmet_0970 GEMM-1b riboswitch are selective for cGAMP and cdiG over a 100-fold range of riboswitch RNA concentration, respectively. (C) LC-MS chromatogram traces for the ion extraction of cGAMP (m/z = 675) from cell lysates for G. sulfurreducens PCA WT or ΔgacA strains. (D) Single nucleotide polymorphisms (SNP) highlighted in red in GSUR3008 that cause cGAMP insensitive variants to demonstrate constitutive expression of pgcA (GSU1761). The C78U and 94 + G variants were isolated in a previous study selecting for increased rate of Fe(III) oxide reduction in G. sulfurreducens (Tremblay et al., 2011). The cGAMP insensitive C78U variant was further confirmed by in-line probing (Kellenberger et al., 2015). The cGAMP insensitive 93 + A variant is described in this study. (E) The GSUR3008 93 + A mutation in G. sulfurreducens eliminates the requirement for GacA or cGAMP for Fe(III) oxide reduction due to the over-expression of pgcA that was confirmed by RNAseq. No other gene expression changes were detected. Raw reads are deposited in the NCBI SRA database PRJNA290373.
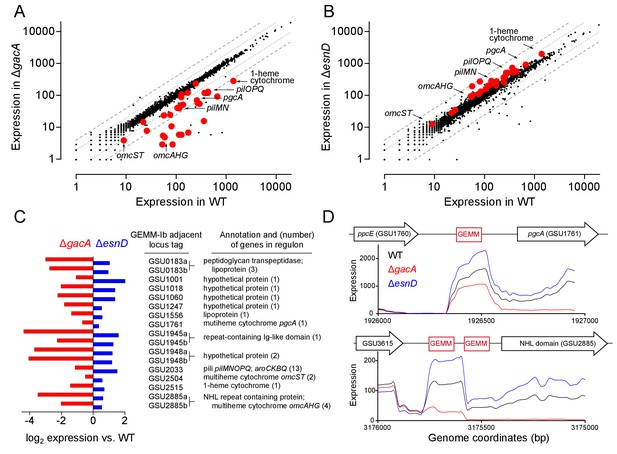
GEMM-1b riboswitch-controlled genes are downregulated in a ΔgacA deletion strain and upregulated in a ΔesnD deletion strain.
Full RNAseq expression data are in Supplementary file 1 (A) Comparison of all genes expressed in WT vs ΔgacA deletion strain. Each dot represents expression of a gene; red dots highlight GEMM-1b regulated genes. Dotted and dashed lines indicate 2- and 4-fold expression differences. Most GEMM-1b regulated genes have at least 2–4 fold decrease in expression compared to WT. (B) Comparison of all genes expressed in WT vs. a ΔesnD deletion strain, labeled as in part A). Most GEMM-1b regulated genes have at least a 2-fold increase in expression compared to WT. (C) GEMM-1b riboswitch transcripts are decreased in a ΔgacA deletion and increased in a ΔesnD deletion compared to WT. GEMM-1b sequences are listed by the first gene locus tag they regulate. Some genes have tandem GEMM-1b sequences upstream, these are labeled by their locus tags followed by ‘a’ and ‘b’. (D) RNAseq reads from WT, ΔgacA, and ΔesnD mapped to the upstream region of GSU1761 (top) and GSU2885 (bottom). GSU2885 is an example of a gene that is regulated by tandem riboswitches. Genes are not drawn to scale with the riboswitch sequences. RNAseq data are generated from biological replicates (n = 2).
-
Figure 3—source data 1
Geobacter mRNA expression levels.
- https://doi.org/10.7554/eLife.43959.007
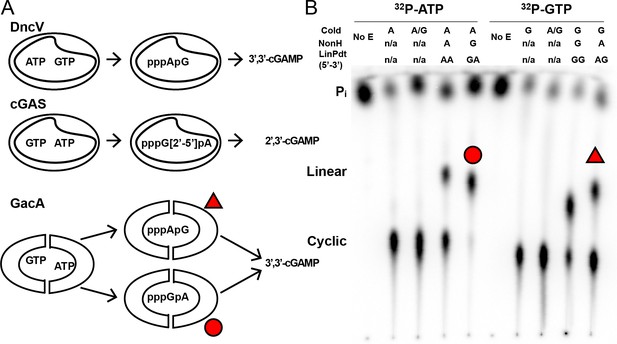
Unlike DncV and cGAS, GacA uses either substrate in the first bond-forming step.
(A) Reaction pathways to form cyclic GMP-AMP by different dinucleotide cyclases, DncV from Vibrio cholerae, cGAS from mammalian cells, and GacA from Geobacter sulfurreducens. (B) Cellulose TLC analysis of radiolabeled products from enzymatic reactions with MBP-tagged GacA R393A (I-site mutant) with NTP substrates and nonhydrolyzable analogues. Trace amounts of α-32P-labeled ATP or α-32P-labeled GTP was doped in the reactions. Reactions were quenched with alkaline phosphatase to digest unreacted nucleotides, resulting in production of inorganic phosphate (Pi).
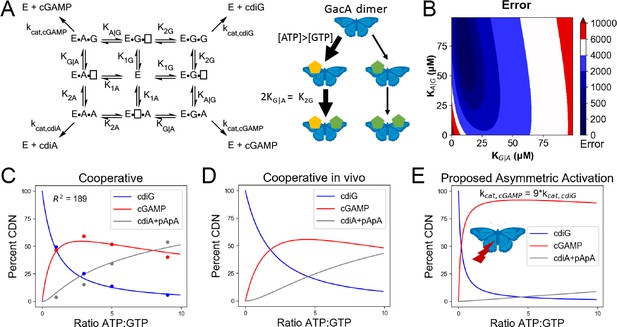
Kinetic analysis of GacA reveals that cooperative binding effects lead to preferential cGAMP production.
(A) Left, reaction pathways for GacA modeled using Python. ‘E’ represents the active enzyme, which is a GacA homodimer. ‘E•N•[ ]’ and ‘E•[ ]•N’ represent enzyme with NTP bound in the first or second half-active sites, respectively, which are treated as equivalent states. ‘E•N•N’ represents enzyme with two NTPs bound. The dissociation constant for the first NTP binding event is K1N, the second binding event is K2N for homodimeric products, and the second binding event is KX|Y for XTP binding after YTP to produce cGAMP. For example, KG|A is the dissociation constant for GTP given GacA already has ATP bound. Right, schematic summarizing kinetic parameters favoring cGAMP production in vivo. (B) Numerical solution of KG|A and KA|G was obtained by varying them between 0–100 µM and minimizing the least squares error (shown) for the modeled product ratios versus the experimental values. The minimum is observed at KG|A = 10 µM and KA|G = 71 µM, giving the best-fit curve shown in (C). (C) Modeled (lines) versus experimental (points) product ratios for GacA at different ATP to GTP ratios. Data points shown are an average of independent technical replicates (n = 3). (D) Modeled product ratios assuming cellular homeostasis of ATP and GTP levels (d[ATP]/dt = d[GTP]/dt = 0). (E) As in (D), except modeled with kcat,AG nine-fold higher than kcat,cdiG or kcat,cdiA. With asymmetric activation, GacA could produce cGAMP almost exclusively.
-
Figure 5—source data 1
GacA product ratios.
- https://doi.org/10.7554/eLife.43959.011
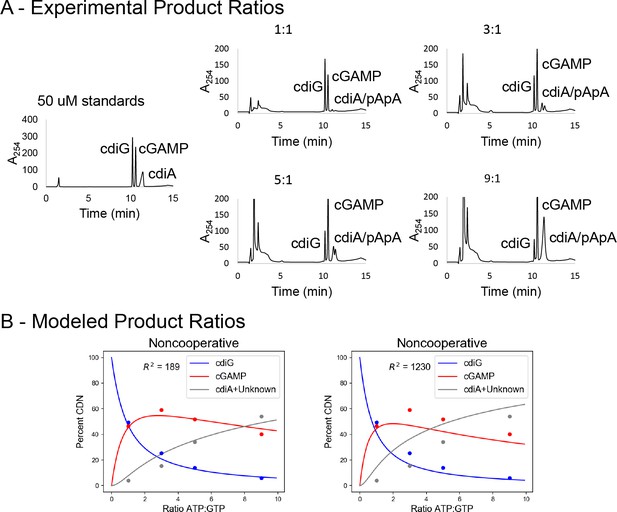
Kinetic analysis of GacA shows better fit for the cooperative model.
Related to Table 1. (A) LC-MS analysis of product ratios for endpoint reactions with 1 µM of 6xHis-MBP-tagged GacA R393A (I-site mutant) using varying ATP:GTP ratios, with [GTP] kept constant at 100 µM. ATP:GTP ratios are shown above each trace. Under these conditions, the cyclic dinucleotides were found to elute in the order of cdiG (10.23 ± 0.02 min), cGAMP (10.56 ± 0.02 min), and cdiA/pApA (11.09 ± 0.05 min). (B) Analysis of product ratios for GacA at different ATP to GTP ratios using Python modeling. Curves represent ratios obtained by fitting to either a noncooperative or cooperative (with KG|A=10 µM, and KA|G=71 µM) mechanism. Data points are an average of n = 3 experiments.
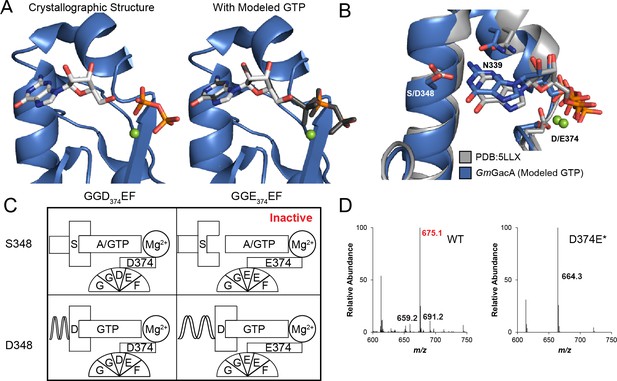
The x-ray crystal structure of GmGacA Hypr GGDEF domain support the role of signature residues and the Goldilocks model.
(A) Active site of the GacA Hypr GGDEF from G. metallireducens (Gmet_1914) with guanosine:PPi bound or modeled GTP based on partial alpha phosphate density. (B) Superposition of x-ray crystal structures of G. metallireducens GacA Hypr GGDEF (blue) and the Idiomarina sp. A28L bacteriophytochrome GGDEF (grey, PDB 5LLX), each with bound guanine nucleotides. Key interacting residues are labeled and shown as sticks. (C) Schematic illustrating the Goldilocks model that explains why the Ser348/GGEEF combination is inactive. In contrast, the flexible Asp side chain at position 348 permits either GGDEF or GGEEF enzymes to remain active. (D) LC/MS analysis of E. coli cell extracts overexpressing G. sulfurreducens GacA WT or D374E* mutant (* indicates that the numbering used corresponds to the G. metallireducens GacA structure, because GsGacA is shorter by one amino acid). Shown are representative MS spectra from integrating the retention time region that would contain the three cyclic dinucleotides. Expected masses are labeled for cdiG (m/z = 691), cGAMP (m/z = 675), and cdiA (m/z = 659). The major peak observed for inactive variants (m/z = 664) is potentially NAD from the lysate.
-
Figure 6—source data 1
LC-MS data for GacA and GGEEF mutant.
- https://doi.org/10.7554/eLife.43959.015
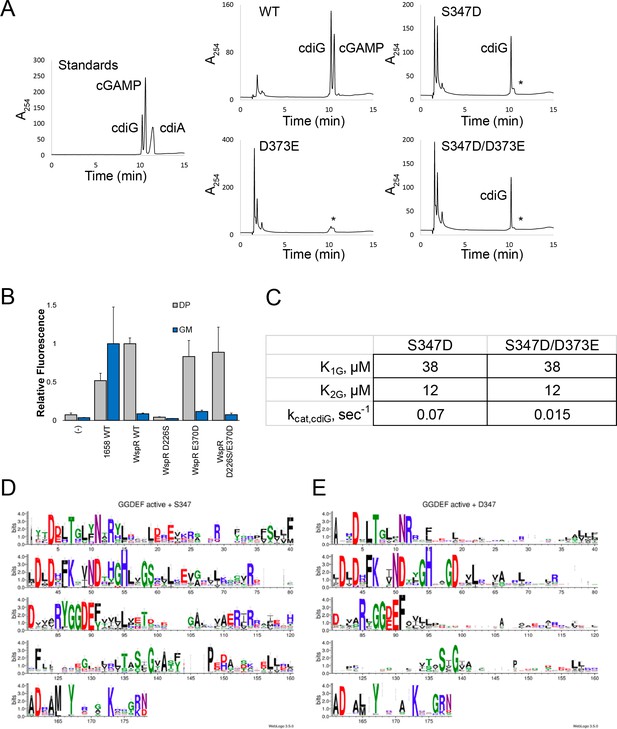
In vitro and in vivo analysis of GacA and WspR mutants to test the ‘Goldilocks’ model.
(A) LC-MS analysis of endpoint reactions with 6xHis-MBP-tagged GacA R393A (I-site mutant = WT) and additional mutants. * represents products that co-elute at similar retention times as cyclic dinuncleotides but that do not have masses assignable to cyclic dinucleotides. (B) Flow cytometry analysis of WspR wildtype and mutants. Average fluorescence measured by flow cytometry (n = 3, 10,000 cells per run) of E. coli BL21 (DE3) Star cells co-expressing the cdiG biosensor Dp-Spinach2 (grey) or cGAMP biosensor GM0970-p1-4delA-Spinach (blue) along with WT GacA (GSU1658), WT WspR, various WspR mutants, or empty vector (-). (C) Kinetic parameters of GacA mutants analyzed in part A for reactions with GTP alone. No significant turnover was observed with ATP alone. GacA R393A D373E was totally inactive with GTP or ATP. (D)-(E) Sequence logo generated from GGDEF domains containing (D) S/T347 (by GSU1658 numbering) or (E) D347. Image generated with WebLogo (Crooks et al., 2004).
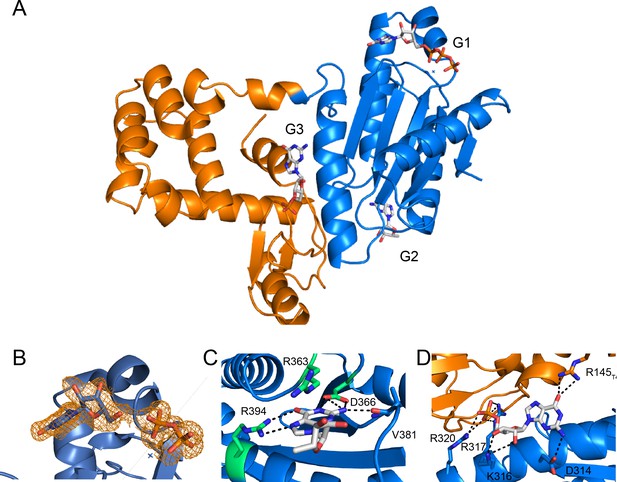
Full x-ray crystal structure shows three guanine nucleotides bound.
(A) Full structure of the T4-Lysozyme-GmetGGDEF fusion showing the location of three guanine nucleotides (G1: active site; G2: I-site; G3: interface). The T4-Lysozyme is in orange and GmetGGDEF is in blue. (B) GTP binding pocket electron density. An Fo-Fc omit map of electron density contoured at 2.0 σ is shown for the bound G1 nucleotide. (C) Interactions of G2 nucleotide bound near the canonical I-site region of the Hypr GGDEF. (D) Interactions of G3 nucleotide at the interface between the T4-Lysozyme and GGDEF domains. The interactions between R145T4 and D314Gmet1914 are expected to occur only for GTP, which may explain our inability to obtain crystals in the presence of ATP.
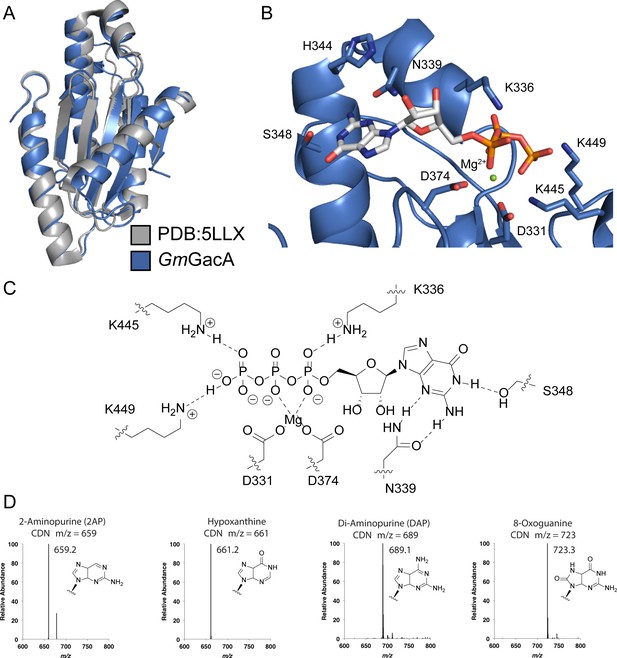
Hypr GGDEF domain structure overlay with canonical GGDEF domain; details of the nucleotide binding pocket.
(A) Overlay of Gmet_1914 GGDEF (blue) with the GGDEF domain of the I. marina light-activated GGDEF (Grey, PDB: 5LLX). (B) Active site of Gmet_1914 GGDEF in complex with GTP. Residues expected to interact with the GTP or Mg2+ cation are labeled. The alpha phosphate electron density was unable to be resolved, but was modeled into the structure. (C) Chemical scheme of active site interactions with GTP substrate. (D) LC-MS analysis of GacA with various unnatural NTP substrates. The structure of the purine base analog and the mass spectra of the homodimeric CDN produced are shown.
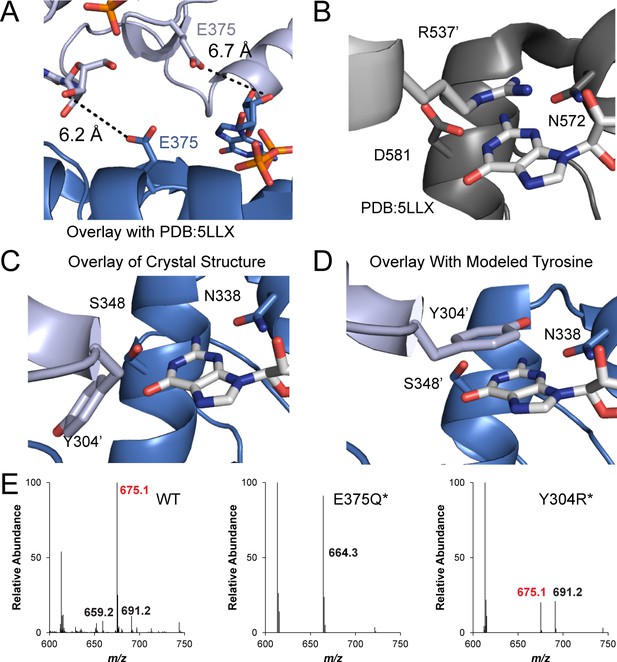
The GmGacA Hypr GGDEF domain structure reveals the molecular basis for hydroxyl activation and substrate-assisted cooperativity for GGDEF enzymes via cross-dimer interactions.
(A) The modeled dimer of GacA GGDEF domains shows that the Glu375 residue from one monomer is poised to deprotonate the 3’-OH of GTP bound in the other half active site. The blue GTP is bound by the blue protein monomer, whereas the grey-blue GTP is bound by the grey-blue protein monomer. (B) and (C) Comparison of the crystallized GGDEF dimer (grey/dark grey, PDB 5LLX) and the modeled GacA dimer (blue/grey-blue) reveals another cross-dimer residue in the half active site that has a different identity between the canonical (D581) and Hypr (Y304’) GGDEFs. (D) Y304' was rotated 180 degrees about the alpha carbon bond from the crystallized structure. (E) LC/MS analysis of E. coli cell extracts overexpressing G. sulfurreducens GacA WT or cross-dimer mutants (* indicates that the numbering used corresponds to the G. metallireducens GacA structure, because GsGacA is shorter by one amino acid). Shown are representative MS spectra from integrating the retention time region that would contain the three cyclic dinucleotides. Expected masses are labeled for cdiG (m/z = 691), cGAMP (m/z = 675), and cdiA (m/z = 659). The major peak observed for inactive variants (m/z = 664) is potentially NAD from the lysate.
-
Figure 9—source data 1
LC-MS data for GacA and cross-dimer mutants.
- https://doi.org/10.7554/eLife.43959.021
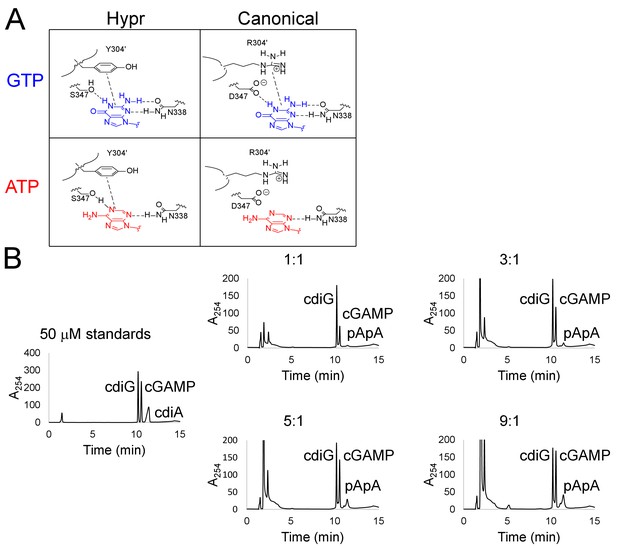
Identification and analysis of a critical cross-dimer interaction with substrate in the active site.
(A) Schematic of the interactions between substrate nucleobases adenine or guanine with either Hypr or canonical GGDEF domains. (B) LC-MS analysis of product ratios for endpoint reactions with 1 µM of 6xHis-MBP-tagged GacA R393A (I-site) and Y303R double mutant using varying ATP:GTP ratios, with [GTP] kept constant at 100 µM. ATP:GTP ratios are shown above each trace.
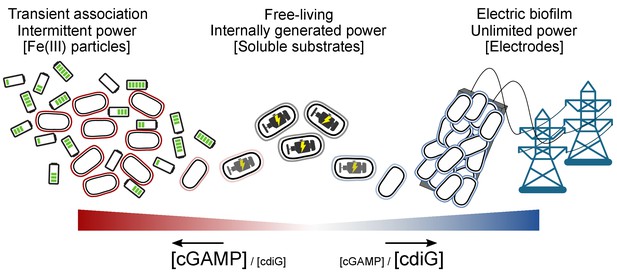
Proposed transient energy lifestyle controlled by cyclic GMP-AMP signaling that is distinct from cyclic di-GMP controlled electrical biofilm.
The second messenger cGAMP is synthesized by GacA, a Hypr GGDEF enzyme, and enhances Fe(III) particle reduction but is not required for growth on electrodes. Cyclic GMP-AMP enhances the transcription of over 30 genes by binding to riboswitches upstream of these genes, including cytochromes and pili. Cyclic di-GMP, commonly used by bacteria for permanently attached lifestyles, is important for growth on electrodes where there is an infinite supply of electron acceptor, but is not required for Fe(III) particle reduction where each particle has a finite electron accepting capacity. To our knowledge, this is the first time that these two contact-dependent electron transfer processes have been shown to be differentially regulated on a global scale.
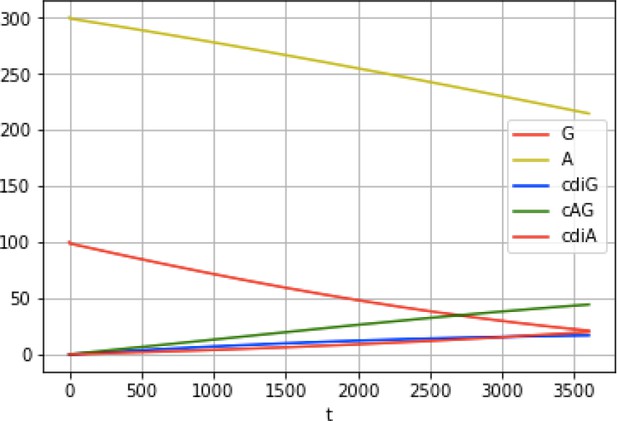
Example reaction time course via simulation.
https://doi.org/10.7554/eLife.43959.023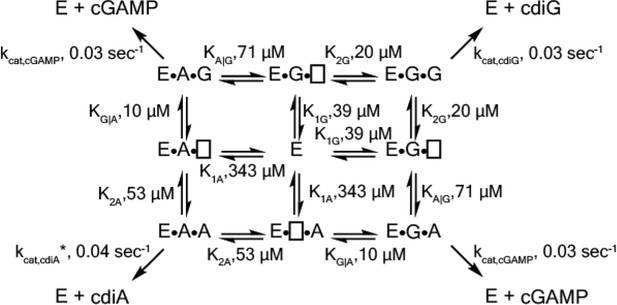
Best fit kinetic model from combined experimental and simulation data.
https://doi.org/10.7554/eLife.43959.024Tables
Kinetic parameters for WT GacA using non-cooperative and cooperative models.
https://doi.org/10.7554/eLife.43959.012| Non-cooperative | Cooperative | |
|---|---|---|
| K1A, µM | 80 | 343 |
| K2A, µM | 80 | 53 |
| kcat,cdiA, sec−1 | 0.04 | 0.03 |
| K1G, µM | 25 | 39 |
| K2G, µM | 25 | 20 |
| kcat,cdiG,sec−1 | 0.04 | 0.03 |
| KA|G, µM | 80 | 71 |
| KG|A, µM | 25 | 10 |
| kcat,cGAMP, sec−1 | 0.04 | 0.03 |
| Model RMSD | 4118 | 443 |
Crystallographic statistics
https://doi.org/10.7554/eLife.43959.018| T4Lys-Gmet_1914 | T4Lys-Gmet_1914 (S Anomalous) | |
|---|---|---|
| Data Collection | ||
| Resolution (Å) | 35.75–1.35 (1.37–1.35) | 48.35–2.72 (2.86–2.72) |
| Wavelength (Å) | 1.11582 | 2.25418 |
| Space group | C 21 | C 21 |
| Unit cell dimensions: a, b, c (Å) | 70.68, 111.65, 55.34 | 71.33, 111.96, 55.66 |
| Unit cell dimensions: α, β, γ (°) | 90, 122.75, 90 | 90.0, 123.10, 90.0 |
| Molecules per ASU | 1 | 1 |
| No. reflections: total | 512201 | 61021 |
| No. reflections: unique | 77765 | 9235 |
| Completeness (%) | 97.8 (67.7) | 94.0 (74.9) |
| Multiplicity | 6.6 (3.1) | 6.6 (5.6) |
| I/σI | 21.6 (2.9) | 21.6 (13.8) |
| CC(1/2)1 (%) | 99.9 (86.2) | 99.4 (99.1) |
| Rpim2 (%) | 1.7 (22.2) | 2.5 (3.2) |
| No. sites | 17 | |
| Refinement | ||
| Resolution (Å) | 35.10–1.35 | |
| Free reflections (%) | 10 | |
| R-factor/R-free | 15.9/17.3 | |
| R.M.S. deviation: bond distances (Å) | 0.016 | |
| R.M.S. deviation: bond angles (°) | 1.560 | |
| Structure/Stereochemistry | ||
| No. atoms: nonhydrogen, protein | 2783 | |
| No. atoms: ligand | 71 (GTP, GMP, PPi) | |
| No. atoms: water | 440 | |
| Average B-factor: nonhydrogen, protein | 18.7 | |
| Average B-factor: ligand | 28.0 | |
| Average B-factor: water | 33.9 | |
| Ramachandran plot: most favored regions | 97.7% | |
| Ramachandran plot: additionally allowed | 2.3% | |
| Protein Data Bank ID | 5VS9 | |
| Reagent type (species) or resource | Designation | Source or reference | Additional information | |
|---|---|---|---|---|
| Strain, strain background (G. sulfurreducens) | Laboratory collection | Cell line maintained in D. bond lab | ||
| Strain, strain background (G. sulfurreducens) | Tn7::cGAMP-nanoluc | this work | Integrated using pCGAMP-9 | |
| Strain, strain background (G. sulfurreducens) | Tn7::cdiG-nanoluc | this work | Integrated using pGGv2-2 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA | this work | Deleted using pDGSU1658 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7:: gacA OE | this work | Integrated using pGSU1658-5 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::gacAWT | this work | Integrated using pGSU1658-6 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::gacAD52A | this work | Integrated using pGSU1658-7 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::gacAR393A | this work | Integrated using pGSU1658-13 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::cAG-nanoluc | this work | Integrated using pCGAMP-9 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::cdiG-nanoluc | this work | Integrated using pGGv2-2 | |
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::cAG- nanoluc / pGacA+ | this work | ||
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::cAG -nanoluc/pGacAWT | this work | ||
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::cAG- nanoluc / pGacAD52A | this work | ||
| Strain, strain background (G. sulfurreducens) | ΔgacA Tn7::cAG- nanoluc / pGacAR393A | this work | ||
| Strain, strain background (G. sulfurreducens) | ΔesnD | Chan et al. (2017) | ||
| Strain, strain background (G. sulfurreducens) | ΔesnD Tn7::esnDOE | this work | Integrated using pGSU3376-4 | |
| Strain, strain background (G. sulfurreducens) | ΔesnD Tn7::cAG-nanoluc | this work | Integrated using pCGAMP-9 | |
| Strain, strain background (G. sulfurreducens) | ΔesnD Tn7::cdiG-nanoluc | this work | Integrated using pGGv2-2 | |
| Strain, strain background (E. coli) | S17-1; recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | Simon et al. (1983) | Donor strain | |
| Strain, strain background (E. coli) | MFDpir; RP4-2-Tc::[ΔMu1::aac(3)IV- ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA | Ferrières et al. (2010) | Donor strain for Tn7 integration | |
| Strain, strain background (E. coli) | MFDpir/pTNS3; MFDpir with plasmid expressing tnsABCD | Choi et al. (2008) | Used for integration downstream of glmS | |
| Strain, strain background (E. coli) | BL21(DE3) star | Life Technologies | ||
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Dp17; pCOLADuet-GSU1658 | Hallberg et al. (2016) | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Gm790p1-4delA; pCOLADuet-GSU1658 | Hallberg et al. (2016) | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Dp17; pCOLADuet-WspR | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Gm790p1-4delA; pCOLADuet-WspR | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Dp17; pCOLADuet-WspR D226S | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Gm790p1-4delA; pCOLADuet-WspR D226S | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Dp17; pCOLADuet-WspR E370D | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Gm790p1-4delA; pCOLADuet-WspR E370D | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Dp17; pCOLADuet-WspR D226S/E370D | this work | For flow cytometry analysis | |
| Strain, strain background (E. coli) | BL21(DE3) star pET31b-Gm790p1-4delA; pCOLADuet-WspR D226S/E370D | this work | For flow cytometry analysis | |
| Recombinant DNA reagent | pRK2-Geo2 | Geobacter expression vector | ||
| Recombinant DNA reagent | pTn7C146 | Tn7 integrative vector, derivative of pTJ1 | ||
| Recombinant DNA reagent | pGSU1658-1 (pGacA+) | GSU1658 (gacA) in pRK2-Geo2 under the control of the acpP promoter | ||
| Recombinant DNA reagent | pGSU1658-8 (pGacAWT) | GSU1658 (gacA) in pRK2-Geo2 under the control of the native gacA promoter | ||
| Recombinant DNA reagent | pGSU1658-9 (pGacAD52A) | GSU1658 (gacAD52A) in pRK2-Geo2 under the control of the native gacA promoter | ||
| Recombinant DNA reagent | pGSU1658-17 (pGacAR393A) | GSU1658 (gacAR393A) in pRK2-Geo2 under the control of the native gacA promoter | ||
| Recombinant DNA reagent | pGSU3376-1 | GSU3376 (esnDOE) in pRK2-Geo2 under the control of the acpP promoter | ||
| Recombinant DNA reagent | pGSU1658-5 | this work | GSU1658 (gacAOE) under the control of the acpP promoter in Tn7 integrative vector | |
| Recombinant DNA reagent | pGSU1658-6 | this work | GSU1658 (gacA) under the control of the native gacA promoter in Tn7 integrative vector | |
| Recombinant DNA reagent | pGSU1658-7 | this work | GSU1658 (gacAD52A) under the control of the native gacA promoter in Tn7 integrative vector | |
| Recombinant DNA reagent | pGSU1658-13 | this work | GSU1658 (gacAR393A) under the control of the native gacA promoter in Tn7 integrative vector | |
| Recombinant DNA reagent | pGSU3376-4 | this work | GSU3376 (esnDOE) under the control of the acpP promoter in Tn7 integrative vector | |
| Recombinant DNA reagent | pK18mobsacB | Simon et al. (1983) | sacB suicide vector for gene deletion | |
| Recombinant DNA reagent | pDGSU1658 | this work | Flanking regions of GSU1658 in pK18mobsacB | |
| Recombinant DNA reagent | pET-MBP-GSU1658 R393A | Hallberg et al. (2016) | Modified pET16a vector containing the GSU1658 R393A mutant with an N-terminal 6xHis-MBP tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET-MBP-GSU1658 S347D/R393A | this work | Modified pET16a vector containing the GSU1658 S347D/R393A mutant with an N-terminal 6xHis-MBP tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET-MBP-GSU1658 D373E/R393A | this work | Modified pET16a vector containing the GSU1658 D373E/R393A mutant with an N-terminal 6xHis-MBP tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET-MBP-GSU1658 S347D/D373E/R393A | this work | Modified pET16a vector containing the GSU1658 S347D/D373E/R393A mutant with an N-terminal 6xHis-MBP tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET24a T4Lysozyme- GmetGGDEF | this work | pET24a vector containing the coding sequence for a chimeric protein consisting an N-terminal T4 lysozyme E11Q mutant followed by residues 294–459 of Gmet_1914 | |
| Recombinant DNA reagent | pET24a GSU1658 | Hallberg et al. (2016) | pET24a vector containing the WT GSU1658 coding sequence with a C-terminal 6xHis tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET24a GSU1658 D373E | this work | pET24a vector containing the GSU1658 D373E coding sequence with a C-terminal 6xHis tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET24a GSU1658 E374Q | this work | pET24a vector containing the GSU1658 E374Q coding sequence with a C-terminal 6xHis tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pET24a GSU1658 Y303R | this work | pET24a vector containing the Y303R GSU1658 coding sequence with a C-terminal 6xHis tag under the control of the T7 promoter | |
| Recombinant DNA reagent | pCOLADuet-1 GSU1658 | Hallberg et al. (2016) | pCOLADuet-1 vector containing the WT GSU1658 coding sequence between the NdeI and XhoI restriction sites. | |
| Recombinant DNA reagent | pCOLADuet-1 WspR | this work | pCOLADuet-1 vector containing the codon-optimized WT WspR coding sequence between the NdeI and XhoI restriction sites. | |
| Recombinant DNA reagent | pCOLADuet-1 WspR D226S | this work | pCOLADuet-1 vector containing the codon-optimized D226S WspR coding sequence between the NdeI and XhoI restriction sites. | |
| Recombinant DNA reagent | pCOLADuet-1 WspR E370D | this work | pCOLADuet-1 vector containing the codon-optimized E370 WspR coding sequence between the NdeI and XhoI restriction sites. | |
| Recombinant DNA reagent | pCOLADuet-1 WspR D226S/E370D | this work | pCOLADuet-1 vector containing the codon-optimized D226S/E370D WspR coding sequence between the NdeI and XhoI restriction sites. | |
| Recombinant DNA reagent | (pCGAMP-1) | this work | The promoter of GSU1761 with cAG selective GEMM-1b riboswitch cloned upstream of nanoluciferase in pTOPO2.1 | |
| Recombinant DNA reagent | (pCGAMP-9) | this work | cAG reporter-nanoluc fusion in pTn7C146, subcloned from pCAG-1 | |
| Recombinant DNA reagent | pGGv2-1 | this work | A cdiG selective variant (A20G) GEMM-1b of Gmet_0970 replaced the GSU1761 GEMM-1b riboswitch cloned upstream of nanoluciferase in pTOPO2.1 | |
| Recombinant DNA reagent | pGGv2-2 | this work | cdiG reporter-nanoluc fusion in pTn7C146 subcloned from pGGv2-1 | |
| Recombinant DNA reagent | pET31b-Gm790p1-4delA | Kellenberger et al. (2015) | pET31b vector expressing the Spinach1-GM790p1-4delA (cAG-selective) biosensor | |
| Recombinant DNA reagent | pET31b-Dp17 | Wang et al. (2016) | pET31b vector expressing the Spinach2-Dp17 (cdiG-selective) biosensor | |
| Sequence-based reagent | Codon-optimized WspR (oligonucleotide) | IDT | ATGCATAATCCGCATGAATCAAA GACGGACCTGGGAGCTCCACTT GACGGAGCCGTGATGGTTTTATT AGTGGACGACCAGGCGATGATCG GTGAGGCGGTCCGCCGTTCTCTG GCTTCTGAAGCGGGCATCGACTTC CATTTTTGCTCCGATCCGCAGCAA GCGGTAGCGGTAGCCAATCAAATT AAGCCCACGGTTATCCTGCAGGAT CTGGTCATGCCTGGCGTGGATGG GCTGACATTGTTAGCAGCTTATCG CGGAAACCCTGCAACACGCGACAT TCCGATCATTGTGCTGAGTACCAA GGAGGAACCCACTGTTAAGTCAGC TGCATTTGCAGCCGGGGCGAATG TGCATTTGCAGCCGGGGCGAATG ACTACCTGGTCAAACTTCCAGATG CGATCGAATTAGTTGCTCGCATCC GCTACCACAGTCGCAGCTACATCG CGCTTCAGCAACGCGATGAAGCCT ACCGCGCCTTGCGCGAATCCCAGC AGCAGCTTCTTGAAACGAACCTGG TTTTGCAGCGTCTGATGAACTCCG ACGGTTTAACGGGTTTGTCTAATC GCCGTCATTTTGATGAATACTTAG AGATGGAATGGCGTCGTAGTTTGC GTGAACAATCTCAGTTGTCATTACT TATGATCGACGTCGACTACTTTAAA TCGTACAACGATACCTTCGGCCATG TAGCGGGTGACGAAGCATTACGTC AAGTCGCTGGCGCGATCCGTGAAGG GTGCTCCCGTTCTTCTGACCTTGCG GCTCGCTATGGTGGAGAGGAGTTTG CAATGGTTCTGCCTGGGACATCACCG GGGGGCGCTCGCCTGTTGGCTGAGA AAGTGCGTCGCACGGTGGAAAGTTTG CAGATCTCGCATGATCAACCGCGTCCA GGCTCGCATTTAACGGTGTCGATCGGC GTATCCACCTTGGTTCCTGGAGGTGGA GGCCAGACCTTTCGCGTTTTGATCGAA ATGGCTGACCAGGCATTATACCAGGCC AAAAATAATGGACGTAATCAGGTGGGA TTGATGGAACAACCAGTACCTCCGGCA CCTGCTGGA | |
Additional files
-
Supplementary file 1
RNAseq.
Raw RNAseq fastq reads from duplicate cultures of each strain was analyzed using Rockhopper (see methods). The re-sequenced and re-annotated reference genome was from our previously published work (Chan et al., 2017). Instead of normalizing the expression to the total number of reads expressed more commonly as reads per kilobase mapped, the expression value is normalized by Rockhopper for each sample to the upper quartile gene expression level, excluding genes with zero expression. This method of normalization better correlates RNAseq expression with qRT-PCR data in a separate test study. q-values are calculated from p-values to control the false discovery rate and used to indicate significance of differentially expression between conditions. A q-value of <0.01 is consider to be significant.
- https://doi.org/10.7554/eLife.43959.025
-
Supplementary file 2
Primers used in this study.
Primers are listed from 5’ - 3’
- https://doi.org/10.7554/eLife.43959.026
-
Transparent reporting form
- https://doi.org/10.7554/eLife.43959.027





