Novel RNA and DNA strand exchange activity of the PALB2 DNA binding domain and its critical role for DNA repair in cells
Figures
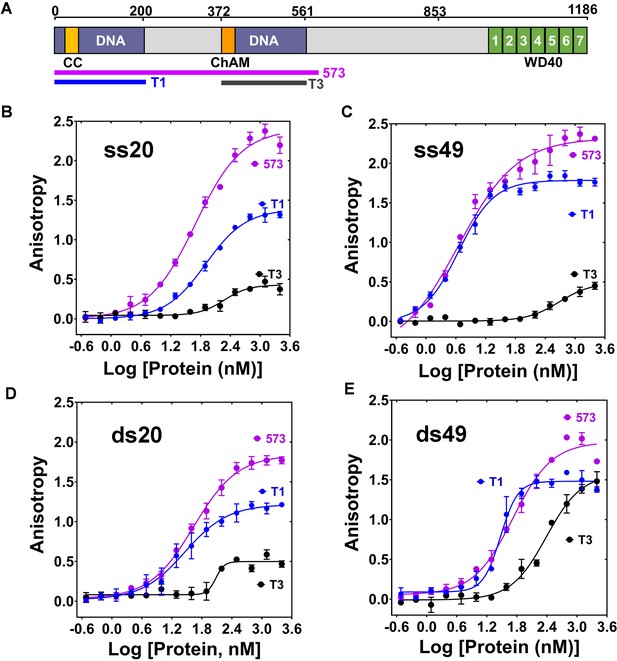
Interaction of T1, T3 and PB2-573 fragments with ss- and dsDNA.
(A) Domain structure of PALB2. The PALB2 truncations used in the present study are shown below by magenta, blue and dark grey lines. (B–E) Equilibrium binding of PALB2 fragments, including T1 (blue), T3 (dark grey) and PB2-573 (magenta), to 20 nt ssDNA (ss20) (B), 49 nt ssDNA (ss49) (C), 20 bp dsDNA (ds20) (D), and 49 bp dsDNA (ds49) (E) monitored by fluorescence anisotropy of FAM-labelled ssDNA (5 nM). Each data point is an average of six readings from two different experiments. Reactions were performed in assay buffer with 20 mM Tris-acetate pH 7.0, 100 mM NaCl, 5% glycerol, 10% DMSO in a 40 μL reaction volume.
-
Figure 1—source data 1
Table with Hill coefficients (n) and equilibrium dissociation constants (Kd, nM) values for graphs in Figure 1B–E.
- https://doi.org/10.7554/eLife.44063.004
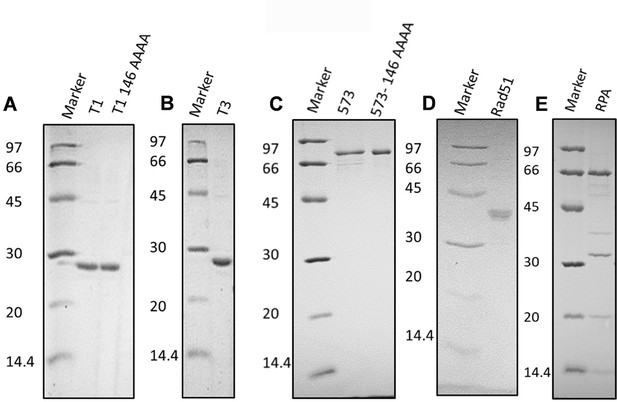
SDS PAGE analysis of purified proteins used in this study: (A) T1 and T1 146AAAA mutant, (B) T3, (C) PB2-573 and PB2-573 146AAAA, (D) RAD51, and (E) RPA.
https://doi.org/10.7554/eLife.44063.005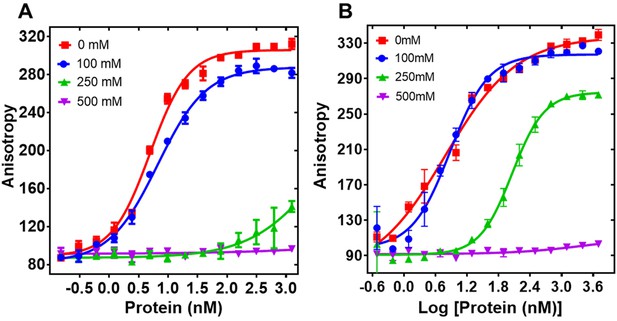
Effect of increasing salt concentration on PALB2 fragments binding to DNA.
(A) Equilibrium binding of T1 to FAM-ss49 at different concentrations of NaCl, (B) equilibrium binding of PB2-573. Each data point is an average of six readings from two different experiments.
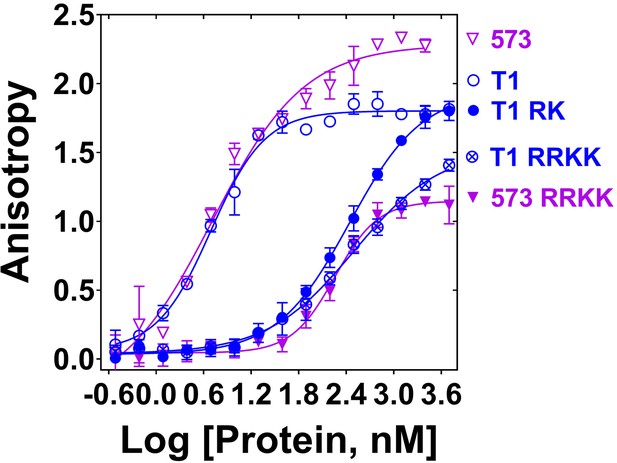
Mutation of DNA binding residues.
Isotherms of fluorescence anisotropy of FAM-ss49 (5 nM) titrated by PALB2 T1 (blue, open circles) and PB2-573 (magenta, open triangles) fragments and their mutants: T1 146-RK/AA (filled blue circles), T1 146-RRKK/AAAA (crossed open blue circles), and 573 146-RRKK/AAAA (filled magenta triangles) under conditions identical to those in Figure 1.
-
Figure 2—source data 1
Table with Hill coefficients (n) and equilibrium dissociation constants (Kd, nM) values.
- https://doi.org/10.7554/eLife.44063.008

Amino acid sequence alignment of PALB2 T1 from different organisms with residues colour-coded accordingly to polarity, with mutated residues identified by red boxes, and with the secondary structure elements depicted at the bottom of alignment in cartoon representation as predicted by the Phyre server.
The following sequences were used in the analysis. gi|21757330 – Homo sapiens, gi|1297718444 - Piliocolobus tephrosceles, gi|724966540 - Rhinopithecus roxellana, gi|967502006 - Macaca mulatta, gi|1060946151 - Callithrix jacchus, gi|1328794872 - Loxodonta africana, gi| 1244140196 - Enhydra lutris kenyoni, gi|821391924 - Orcinus orca, gi|1111220295 - Panthera pardus, gi|664756985 - Equus przewalskii, gi|124486979 - Mus musculus.
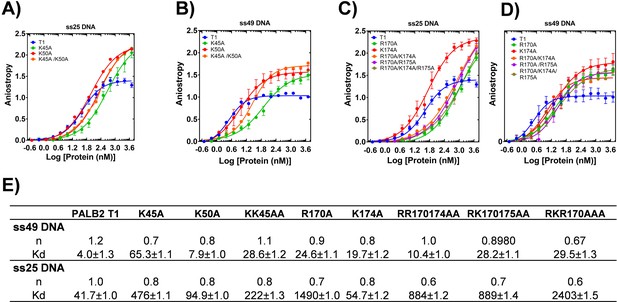
DNA binding of T1 mutants.
Equilibrium binding of 5 nm FAM-ss25 (A, C) or FAM-ss49 (B, D) to PALB2 N-DBD and its mutants. (blue) In A) and B) isotherm for K45A is in green, for K50A in red, for K45A/K50A in black. In C) and D) isotherm for R170A is in red, for K174A in green, for R170A/K174A in magenta, for R170A/R175A in orange, and for R170A/K174A/R175A in black. The binding buffer is 20 mM Tris-acetate pH 7.0, 100 mM NaCl, 5% glycerol, 10% DMSO.
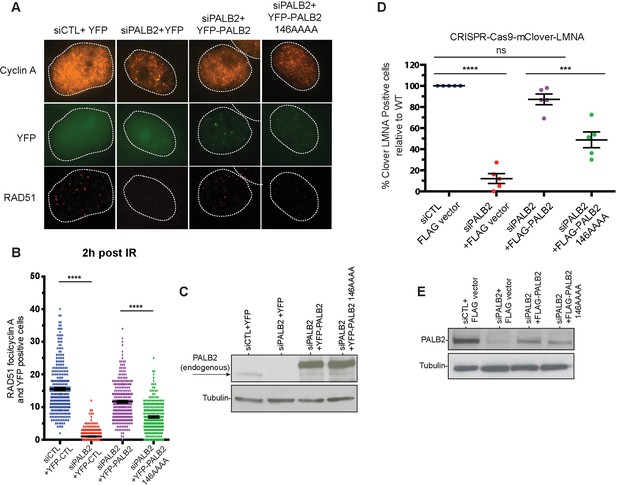
Effect of a PALB2 DNA-binding site mutation on homologous recombination.
(A) Representative immunofluorescence images of RAD51 foci in PALB2 knockdown HeLa cells complemented with the indicated YFP construct and synchronized in S/G2 by double thymidine block, as determined by cyclin A staining. (B) RAD51 foci quantification in control siRNA (blue), siPALB2 (red) and with siPALB2 with subsequent complementation by siRNA-resistant constructs YFP-PALB2 (magenta) and 146AAAA DNA-binding site mutant PALB2 (green) at 2 hr after irradiation. (C) Western blotting of the samples shown in (B) to monitor knockdown and complementation efficiency. (D) Quantification of the gene-targeting efficiency of siRNA PALB2 cells complemented with wild-type and 146AAAA siRNA-resistant constructs mClover positive/iRFP cells. (E) Western blotting of the samples shown in (D) to monitor knockdown and complementation efficiency. ***p<0.001 and ****p<0.0001.

Time dependence of RAD51 foci formation after DNA damage in the experiment described in Figure 3.
Cells were prepared as described in Figure 3 and processed for immunofluorescence at 1 hr, 2 hr, 4 hr, 6 hr and 8 hr post-irradiation.

Representative images and schematic representation of CRISPR-Cas9/mClover-LMNA1 mediated HR assay.
Following nucleofection, Cas9 creates a double-strand break in the LMNA locus leading to integration of the mClover gene at the LMNA locus by homologous recombination. Clover-labelled Lamin A/C proteins exhibit green fluorescence enriched at the nuclear periphery, which is indicative of successful gene targeting by homologous recombination.
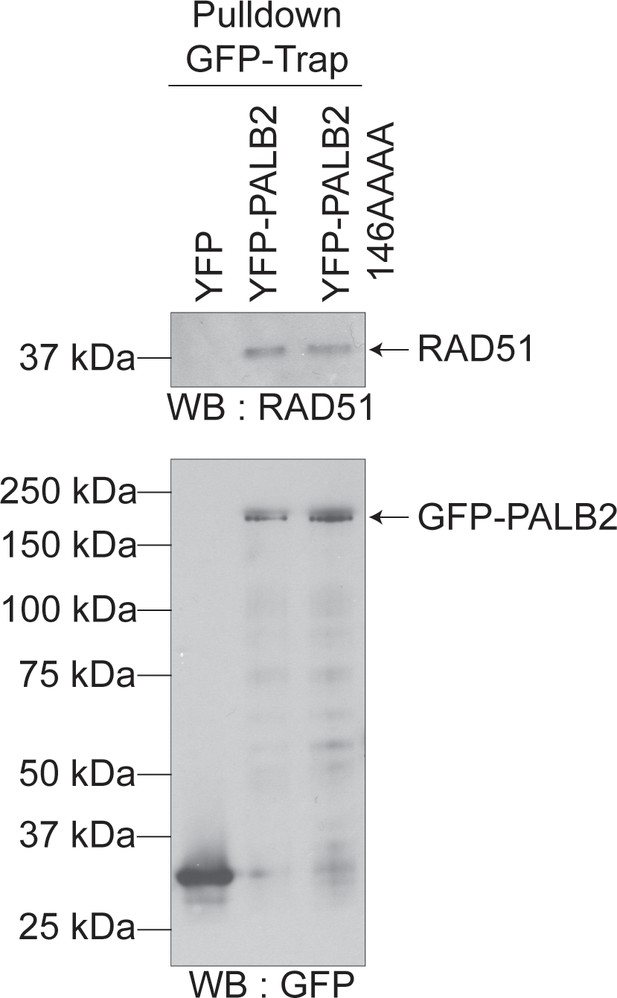
YFP-PALB2 and YFP-PALB2 146AAAA binds RAD51 equally.
Lysates from HEK293T cells expressing the indicated constructs were subjected to GFP-Trap pulldown and immunoblotting against YFP or RAD51. YFP alone was used as a negative control.
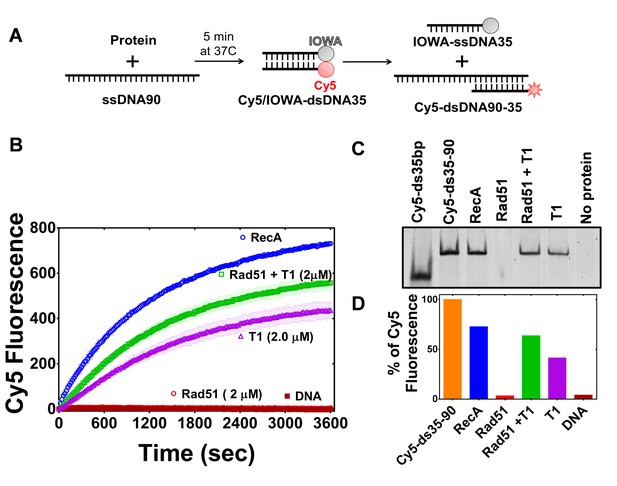
PALB2 promotes strand exchange between homologous DNA substrates.
(A) Schematic representation of the strand exchange activity assay. ss90 (120 nM) was incubated with RecA (2 μM) or RAD51 (2 μM) for 5’, then with PALB2 fragment (2 μM) for 5’, then dsDNA (100 nM) was added and the Cy5 fluorescence was measured on a plate reader using 635 nm excitation and 680 nm emission wave-lengths. (B) Continuously measured Cy5 fluorescence after initiating reactions with RecA (blue), RAD51 (red), PALB2 N-DBD (magenta), RAD51 and PALB2 N-DBD (green), and without proteins (dark red). Each point is an average of three measurements. (C) Reaction products from (B) were deproteinized and separated on a native PAGE gel. Control in lane one represent Cy5-labelled dsDNA without IOWA quencher and in lane 2 Cy5-labelled ssDNA annealed with ss90. (D) Percentage of Cy5 fluorescence of the final reaction in (B).

RAD51 activity under optimized conditions.
(A) Strand exchange by RAD51 monitored by Cy5 fluorescence under optimized conditions with 5 mM CaCl2 and 5 mM MgCl2. (B) End products of reactions in (A) analyzed with EMSA.
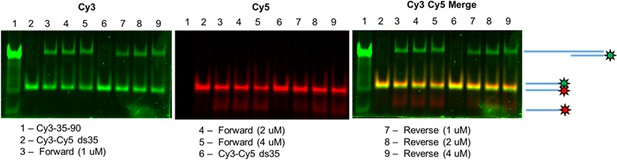
PALB2 strand exchange activity with dsDNA with alternative fluorophores.
EMSA of N-DBD-mediated strand exchange products using dsDNA composed of Cy3- and Cy5-labelled DNA strands. The reaction was performed identically to the one in Figure 4, but without ATP in buffer. The end products of the reaction were deproteinized, separated by EMSA and imaged with a Typhoon system using Cy3 signal (left panel) and Cy5 signal (middle panel). The right panel represents an overlap of left and middle panels. The lanes are 1) control of Cy3-labaled oligo annealed to ss90, 2) Cy3/Cy5 dsDNA, 3), 4), 5) end product of forward strand exchange reaction (see Figure 7 below) performed with 1, 2 and 4 μM N-DBD, 6), 7), 8) end products of the reverse strand exchange performed with 1, 2, and 4 μM N-DBD.
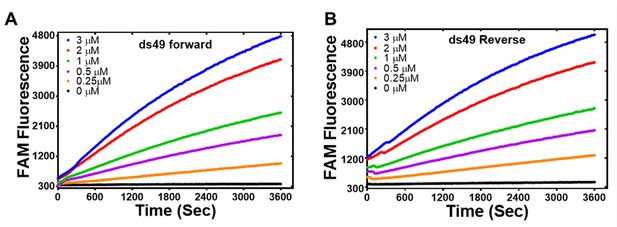
PALB2 strand exchange activity with longer dsDNA.
Forward (A) and inverse (B) strand exchange reactions supported by N-DBD at different concentrations ranging from 0.25 μM to 3 μM performed with ss90 and FAM/Dadsyl-labelled ds49.

PALB2 N-DBD cannot form D-loops with supercoiled dsDNA plasmid.
Products of the D-loop reaction mediated by PALB2 N-DBD fragment with 100-mer single strand DNA oligonucleotide (JYM1413, 1 µM) and supercoiled dsDNA plasmid pPB4.3 (300 µM) for 5 min separated by gel electrophoresis. RAD51 (1 µM) was used as a positive control (line 2). ‘ccc’ line is a control without plasmid. T1 fragment did not stimulate RAD51 D-loop formation activity in this assay as well (not shown).
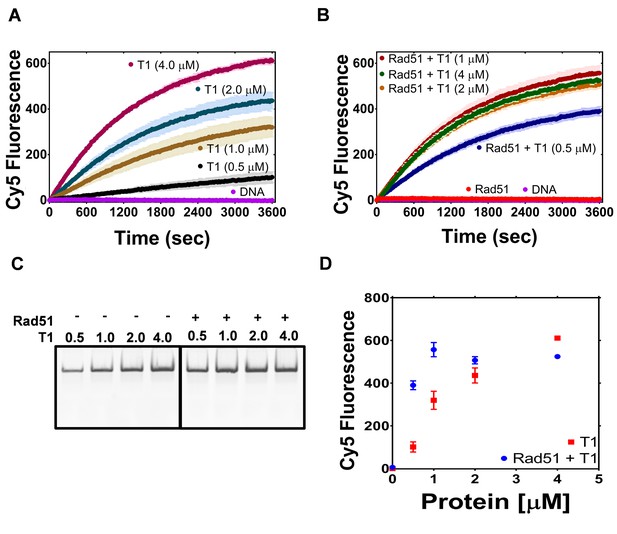
The PALB2 N-DBD stimulates RAD51 strand exchange.
(A) Dependence of the strand exchange activity on N-DBD concentration: 0.5 μM (black), 1 μM (brown), 2 μM (navy) and 4 μM (dark red). (B) A reaction similar to (A) in the presence of RAD51 (2 μM, red: without N-DBD). Each point is an average of three measurements in (A) and (B). (C) Deproteinated strand exchange activity products from (A) and (B) separated on native PAGE gel. (D) End-point values of the strand exchange reactions shown in (A) and (B) plotted against N-DBD concentration. Buffer and DNA concentration are identical to those in Figure 4.
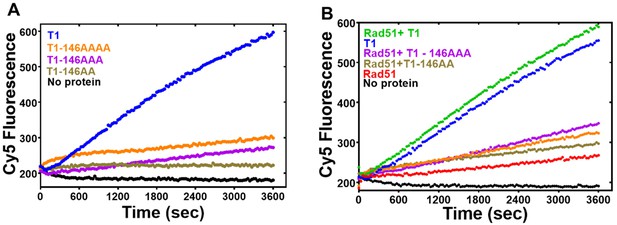
PALB2 DNA binding site mutants do not support strand exchange.
(A) Forward strand exchange activity of PALB2 N-DBD DNA binding mutants. (B) The strand exchange activity of PALB2 N-DBD mutants in the presence of Rad51. Strand exchange reactions were performed with Cy5 and Iowa labelled 35 bp DNA at 2 μM protein concentration.
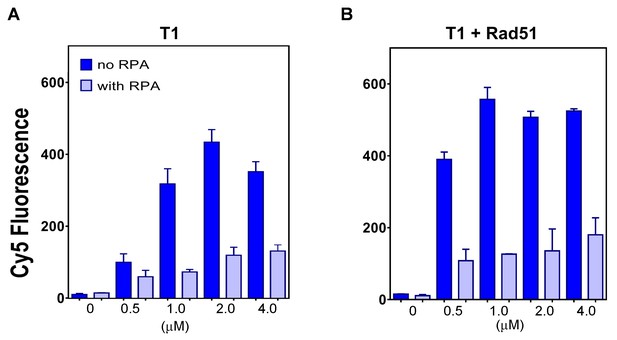
The plot of end points of strand exchange reactions with and without 0.5 μM RPA by (A) N-DBD and (B) N-DBD in the presence of 2 μM RAD51.
Reactions are performed identically to those in Figure 5.
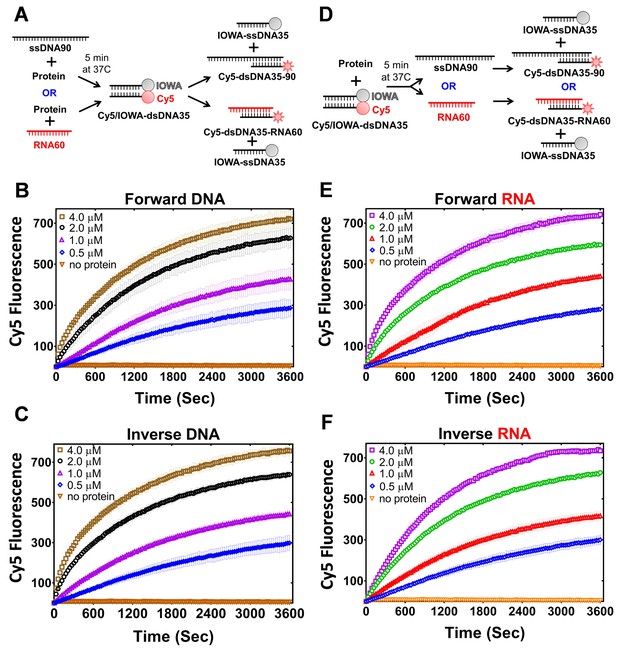
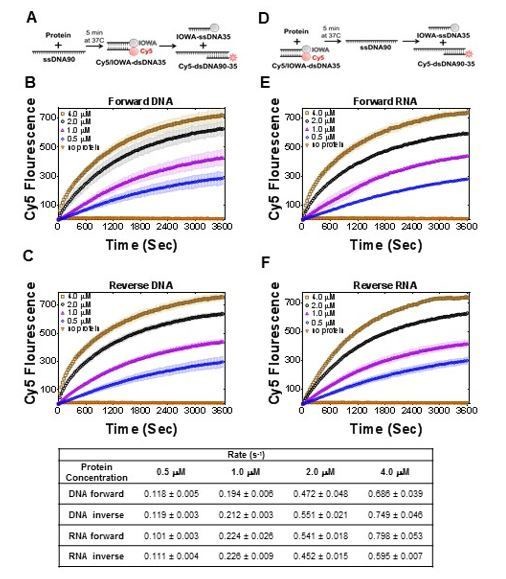
PALB2 promotes forward and inverse strand exchange with ssDNA and RNA substrates.
Schematic representation of forward (A) and inverse (D) reactions. Cy5 fluorescence change for the forward reaction with ssDNA is shown in (B) and with RNA in (E) at different concentrations of T1 fragment ranging from 4 μM (brown in B, magenta in E) to 0.5 μM (blue) and without protein in orange. Similar time courses of inverse reactions for ssDNA are shown in (C) and for RNA in (F). Buffer and DNA concentration are identical to those in Figure 4. Each point is an average of three measurements.
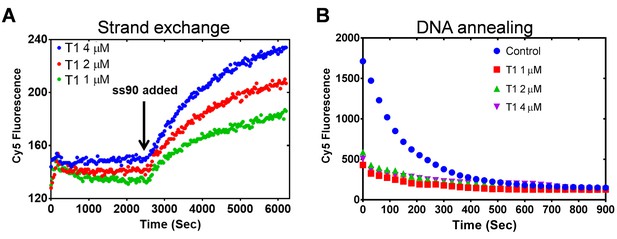
PALB2 does not unwind dsDNA and anneals complementary DNA strands.
(A) Strand exchange reaction where Cy5/Iowa ds35 DNA (100 nM) was first incubated with three different concentrations of N-DBD for 30’. The complementary ss90 DNA (100 nM) was added at 30' to initiate the strand exchange. (B) Annealing of Cy5- and Iowa-labelled complementary ss35 strands (100 nM) in the presence of different concentrations of PALB2 N-DBD. Buffers are identical to those in Figure 4.
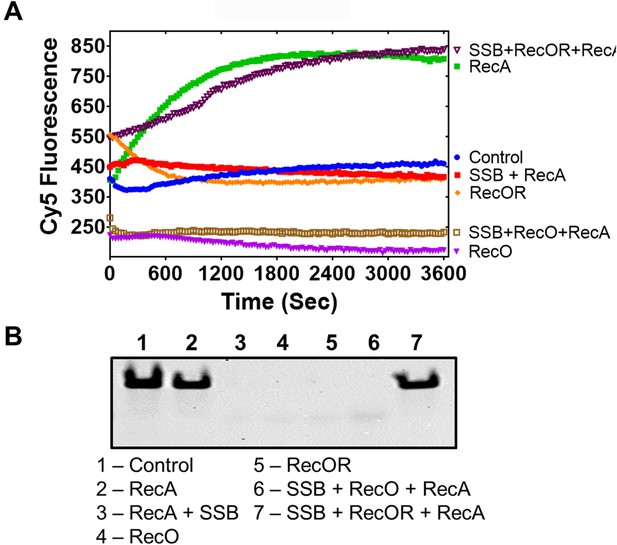
RecO and RecOR do not support strand exchange without RecA.
(A) Strand exchange activity of RecO (magenta), RecOR (orange), RecA (green), RecA with SSB (red), RecA with RecOR and SSB (dark brown), and RecO with RecA and SSB (brown) measured under conditions identical to those in Figure 4 monitored by Cy5 fluorescence. (B) Reaction products from A) were deproteinized and separated on native PAGE gel.
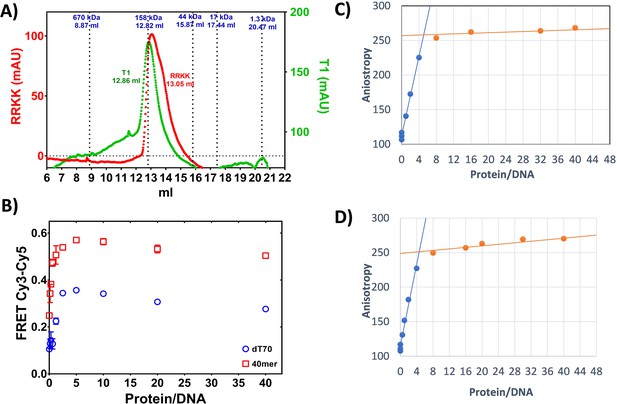
Oligomerization and DNA-binding stoichiometry of PALB2 N-DBD.
(A) Gel filtration of the N-DBD (blue isotherm) and the 146AAAA mutant (red) using Superdex-200 10/300 column. Positions of BioRad MW standards with MW in kDa are shown on top. The elution volume of N-DBD correspond to tetrameric or pentameric structure. (B) FRET values of Cy3-ss40-Cy5 (red) and Cy3-dT70-Cy5 (blue) oligonucleotides upon titration by N-DBD. (C) and D) Titration of FAM-labelled ss49 at 50 nM in (C) and 100 nM in (D) by N-DBD.
DNA and T1 were incubated at 37C for 30 min and treated with proteinase K for 20 min and ran on Native page gel at 60 V for 30 min.
DNA without protein were treated exactly the same and compared on rhe gel.
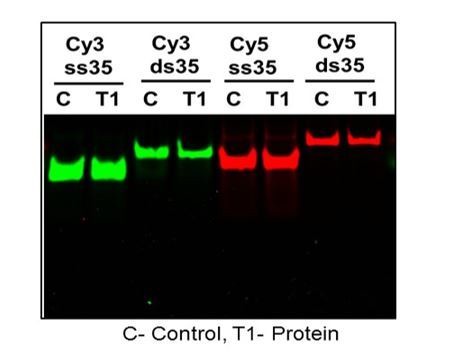
Table with initial rates (bottom) calculated for reactions in Figure 7.
https://doi.org/10.7554/eLife.44063.030Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Human) | HeLa | ATCC | CCL-2 | |
| Cell line (Human) | U2OS | ATCC | HTB-96 | |
| Cell line (Human) | HEK293T | ATCC | CRL-11268 | |
| Sequence-base reagent | siRNA sequences | This paper | Supplementary file 1E | |
| Sequence-base reagent | oligonucleotide JYM1413 | This paper | Supplementary file 1C | |
| Chemical compound, drug | Lipofectamine RNAiMAX | Invitrogen | 13778–150 | |
| Chemical compound, drug | Thymidine | Sigma | T1895-1G | |
| Chemical compound, drug | Lipofectamine 2000 | Invitrogen | 11668019 | |
| Chemical compound, drug | Proteinase K | Sigma | P2308-100MG | |
| Chemical compound, drug | [gamma-32P] ATP | Perkin Elmer | NEG502A250UC | |
| Recombinant DNA reagent | YFP-CTL or YFP (plasmid) | Pauty et al., 2017 | ||
| Recombinant DNA reagent | YFP-PALB2 (plasmid) | Pauty et al., 2017 | ||
| Recombinant DNA reagent | YFP-PALB2 146AAAA (plasmid) | This paper | Supplementary file 1B | |
| Recombinant DNA reagent | pCR2.1-mClover- LMNAdonor (plasmid) | Pinder et al., 2015 | ||
| Recombinant DNA reagent | pX330-LMNAgRNA (plasmid) | Pinder et al., 2015 | ||
| Recombinant DNA reagent | iRFP670 (plasmid) | Pinder et al., 2015 | ||
| Recombinant DNA reagent | FLAG vector (plasmid) | Pauty et al., 2017 | ||
| Recombinant DNA reagent | FLAG-PALB2 (plasmid) | Pauty et al., 2017 | ||
| Recombinant DNA reagent | FLAG-PALB2 146AAAA (plasmid) | This paper | Supplementary file 1B | |
| Recombinant DNA reagent | pPB4.3 | PMID:15899844 | ||
| Other | DAPI stain | Invitrogen | D1306 | (1 µg/mL) |
| Commercial assay, kit | SE Cell Line 4D-Nucleofector X Kit | VWR | CA10064-148 | |
| Antibody | anti-RAD51 (mouse monoclonal) | Novus Biologicals | NB100-148 | (1:1000) |
| Antibody | anti-GFP (mouse monolconal) | Roche | 11814460001 | (1:1000) |
| Antibody | RAD51 (rabbit polyclonall) | B-Bridge International | 70–001 | (1:7000) |
| Antibody | cyclin A (mouse monoclonal) | BD Biosciences | 611268 | (1:400) |
| Antibody | Alexa Fluor 568 goat anti-rabbit | Molecular Probe | A11011 | (1:10000) |
| Antibody | Alexa Fluor 647 got anti-mouse | Molecular Probe | A21235 | (1:10000) |
| Software, algorithm | Prism | GraphPad | Ver-6 | |
| Software, algorithm | Volocity | Quorum Technologies | v6.0.1 | |
| Sequence-base reagent | cloning primers, DNA-binding substrates, FRET substrates | IDT, Technology | Supplementary file 1B,C,D | |
| Strain, Strain background (E. Coli) | BL21-star cells | ThermoFisher Scientific | C601003 | |
| Strain, Strain background (E. Coli) | OmniMAX cells | ThermoFisher Scientific | C8540-03 | |
| Recombinant DNA reagent | pSMT3-PALB2-T1 | This paper | ||
| Recombinant DNA reagent | pSMT3-PALB2-T3 | This paper | ||
| Recombinant DNA reagent | pSMT3-PALB2-573 | This paper | ||
| Other | His60 Ni Superflow | Clonetech | 635660 | |
| Other | Heparin 5 ml HiTrap column | GE Heathcare Lifesciences | 17040701 | |
| Other | Superdex-200 10/300 GL column | GE Heathcare Lifesciences | 17517501 | |
| Recombinant DNA reagent | pET11-Rad51 | Dr. A Mazin Lab; PMID:11751636 | ||
| Recombinant DNA reagent | pSMT3-RAD51 | This paper | ||
| Recombinant DNA reagent | p11d-tRPA | AddGene, Henricksen et al., 1994 | 102613 | |
| Other | Affi-Gel blue affinity gel | BioRad | 1537301 | |
| Instrument | Synergy four plate reader | BioTek |
Additional files
-
Supplementary file 1
Supplementary tables including.
(A) DNA binding parameters of N-DBD mutants, and DNA/RNA sequences for (B) primers for DNA binding site mutagenesis, (C) substrates for DNA binding assay, (D) Substrates for strand exchange activity and FRET assays, (E) siRNA resistance primers.
- https://doi.org/10.7554/eLife.44063.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44063.028





