RETRACTED: Pyrophosphate modulates plant stress responses via SUMOylation
Figures
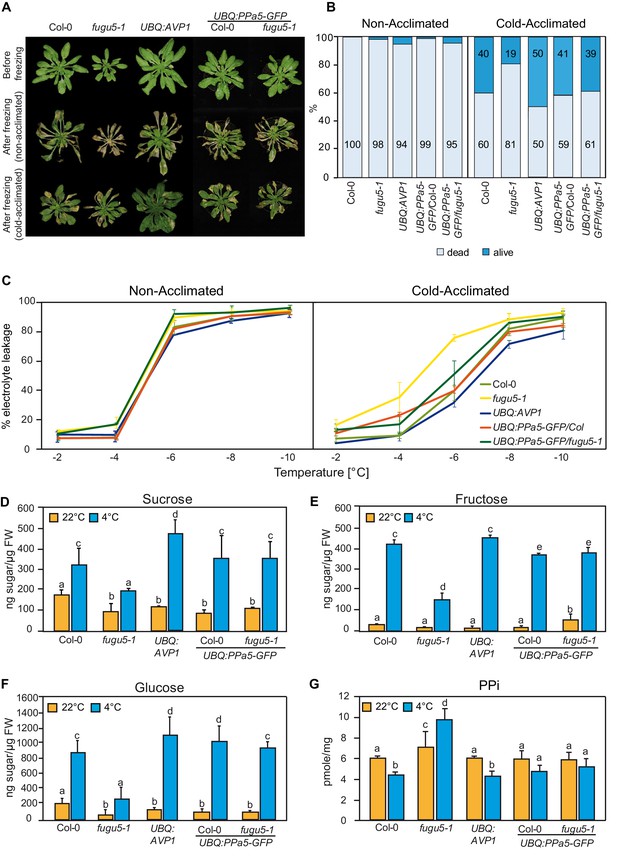
PPi hydrolysis is required to rescue the freezing sensitive phenotype of fugu5-1.
(A) and (B) Freezing tolerance assay. Col-0, fugu5-1, UBQ:AVP1 and UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds were grown for 6 weeks at 22°C and were then moved to 4°C for cold-acclimation, or kept at 22°C for 4 days. Afterwards, plants were subjected to a 5 hr freezing temperature regime (0 to −10°C). After thawing at 4°C overnight, plants were moved back to 22°C. (A) Images were taken before cold-acclimation and one week after the freezing treatment. (B) Quantification of dead and alive leaves was done one week after the freezing treatment with n ≥ 75 leaves from 5 individual plants. 3 independent experiments were performed. Cold-acclimated UBQ:AVP1 and fugu5-1 are significantly different compared to the other genotypes (Student’s t-test p<0.05). (C) Electrolyte leakage assay of Col-0, fugu5-1, UBQ:AVP1 and UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds was performed on leaf material of acclimated and non-acclimated plants at indicated freezing temperatures. Error bars represent SD of the mean of n = 3 biological replicates. (D–G) Sugar and PPi measurements were done from extracts of acclimated (4°C) and non-acclimated (22°C) 6-week-old rosette leaves. Error bars show SD of the mean with n = 3 samples of one representative experiments. 3 biological replicates were performed. Significant differences are indicated by different letters (Two-way ANOVA followed by Tukey’s test, p<0.05).
-
Figure 1—source data 1
(B) Survival of the 6 weeks old rosette leaves upon freezing.
(C) Electrolyte leakage analysis of the cold-acclimated and non-acclimated 6-weeks old rosette leaves upon exposure to freezing temperature. (D-G) Sugar and PPi measurements of the 6-weeks old rosette leaves.
- https://doi.org/10.7554/eLife.44213.008
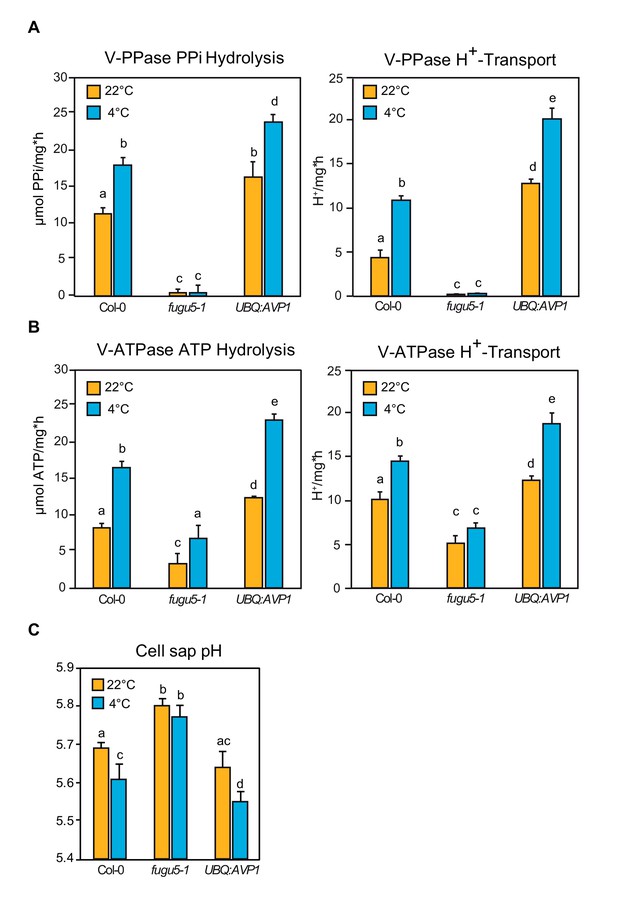
Increased Proton Transport Activity of V-ATPase upon Cold Acclimation Requires an Active V-PPase.
(A) Enriched tonoplast proteins were used to determine K+-stimulated PPi hydrolysis and H+ transport activity of V-PPase. (B) Enriched tonoplast proteins were used to determine KNO3-inhibited ATP hydrolysis and H+ transport activity of V-ATPase. (C) Cell sap pH measurement of rosette leaves. Error bars show SD of the mean with n = 12 samples of 3 biological replicates. (A–C) Col-0, fugu5-1 and UBQ:AVP1 were grown for 6 weeks under short-day conditions then were cold acclimated for 4 days at 4°C. Untreated plants were maintained in the same conditions as the growth period. Error bars represent SD of n = 3 biological replicates. Significant differences are indicated by different letters (Two-way ANOVA followed by Tukey’s test, p<0.05).
-
Figure 1—figure supplement 1—source data 1
(A) V-PPase PPi hydrolysis and H+- transport measurements.
(B) V-ATPase ATP hydrolysis and H+-transport measurements. (C) Cell sap pH measurements.
- https://doi.org/10.7554/eLife.44213.004
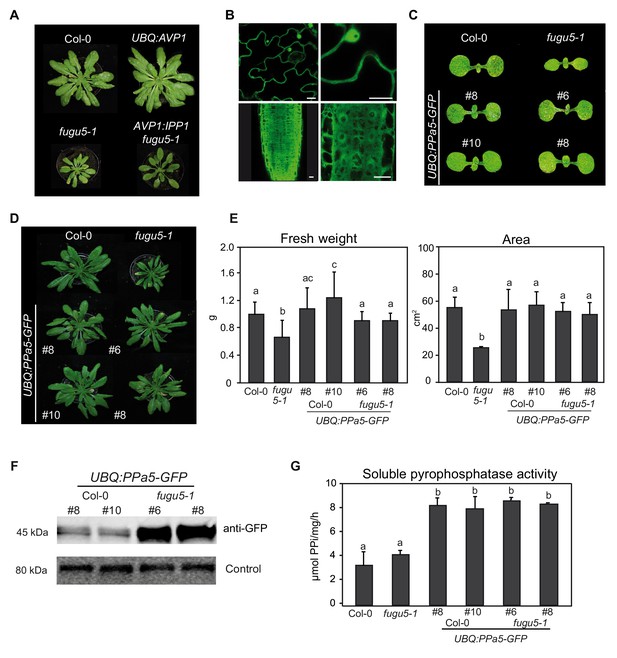
Overexpression of the Arabidopsis soluble pyrophosphatase PPa5 is sufficient to complement fugu5-1.
(A) Comparison of 6-week-old rosettes phenotypes of Col-0, fugu5-1 and overexpression lines of Arabidopsis vacuolar pyrophosphatase AVP1 and yeast soluble pyrophosphatase IPP1. (B) Representative images showing the localization of UBQ:PPa5-GFP to cytosol and nucleus in shoot (upper panel) and root cells (lower panel). Scale bars: 10 µm. (C) Cotyledon phenotypes of 5 days old seedlings of Col-0, fugu5-1 and UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds. (D) Comparison of 6-week-old short day grown rosette phenotypes of Col-0, fugu5-1 and UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds. (E) Measurements of fresh weight and whole rosette area of Col-0, fugu5-1 and UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds. Plants were grown for 6 weeks under short-day conditions. To determine rosette area Rosette Tracker plug-in of ImageJ is used. Error bars represent SD of the mean of n = 20 of 3 biological replicates. (F) Analysis of UBQ:PPa5-GFP protein amount in Col-0 and fugu5-1 background with anti-GFP. Soluble proteins from 6-week-old rosettes grown under short-day conditions were extracted. An internal control provided by SPL detection kit (DyeAGNOSTICS) was used for normalization. One representative image from three biological replicates is depicted. (G) Soluble proteins of Col-0, fugu5-1 and UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds were used to determine K+-stimulated PPi hydrolysis. Plants were grown for 6 weeks under short-day conditions. Error bars represent SD of n = 3 biological replicates. Significant differences are indicated by different letters (Two-way ANOVA followed by Tukey’s test, p<0.05).
-
Figure 1—figure supplement 2—source data 1
(E) Fresh weight and area measurements of the 6-weeks old rosette leaves.
(G) Soluble pyrophosphatase activity measurements.
- https://doi.org/10.7554/eLife.44213.006
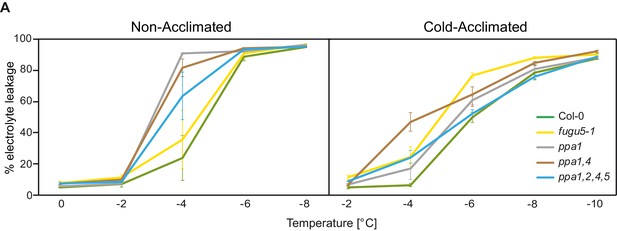
Freezing tolerance phenotypes of the sPPase mutants.
(A) Wt, fugu5-1, ppa1, ppa1,4, ppa1,2,4,5 were grown for 6 weeks at 22°C and were then moved to 4°C for cold-acclimation, or kept at 22°C for 4 days. Electrolyte leakage was performed on leaf material of acclimated and non-acclimated plants at indicated temperatures. Error bars represent SD of the mean of n = 2 independent experiments.
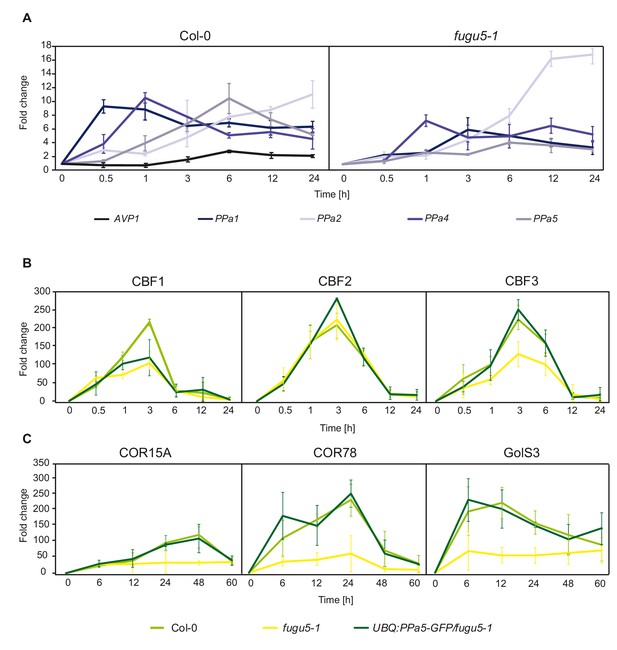
sPPase expression is induced upon cold exposure to control PPi that affects expression of CBFs and CBF target genes.
(A) qRT-PCR for the analysis of expression of PPa 1,2,4,5 and AVP1 in Col-0 and PPa 1,2,4 and 5 in fugu5-1. (B-C) Measurement of expression of CBF, COR and GolS3 genes in Col-0, fugu5-1 and UBQ:PPa5-GFP/fugu5-1 by qRT-PCR. (A-C) and () Plants were grown for six weeks under short-day conditions at 22°C. Afterwards, they were exposed to 4°C for indicated time periods. Whole rosettes were used for total RNA extraction. Actin2 expression was used for normalization. Error bars represent SD of the mean of n = 3 biological replicates. Data analysis was performed using the ΔΔCt method.
-
Figure 2—source data 1
(A) Relative expression of the vacuolar and soluble pyrophosphatases in wt and the relative expression of the soluble pyrophosphtases in fugu5-1 exposed to 4°C for different hours.
(B-C) Relative expression of the cold regulated genes in wt, fugu5-1 and UBQ:PPa5-GFP/fugu5-1 upon exposure to 4°C for different hours.
- https://doi.org/10.7554/eLife.44213.010
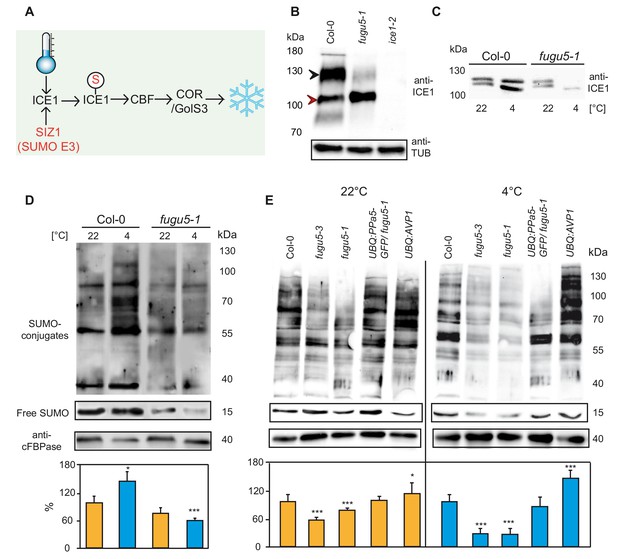
Modification of ICE1 and general SUMOylation upon cold exposure are inhibited in fugu5.
(A) Cold acclimation induces ICE1 SUMOylation which then activates CBFs and leads to the expression of downstream targets for the establishment of freezing tolerance. (B) Determination of the amount of ICE1 in Col-0, fugu5-1 and ice1-2 seedlings. Amount of TUBULIN was detected as loading control. Plants grown in long day conditions for 3 weeks. Black arrow indicates modified ICE1 dimer whereas red arrow indicates unmodified ICE1 dimer. (C) Comparison of the amount of the ICE1 under normal conditions (22°C) and after cold treatment (4°C, 3 hr) in Col-0 and fugu5-1 seedlings. 10-days-old liquid grown seedlings were used for total protein extraction. Anti-ICE1 was used as primary antibody. (D) Western blots comparing SUMOylation levels of Col-0 and fugu5-1 under normal conditions (22°C) and after cold treatment (4°C, 3 hr). (E) Western blots demonstrating the total SUMOylation in Col-0, fugu5-1, fugu5-3, UBQ:AVP1 and UBQ:PPa5-GFP under normal conditions (22°C) and after cold treatment (4°C, 3 hr). (D) and (E) 10 days old liquid grown seedlings were used for total protein extraction. Anti-SUMO1/2 was used as primary antibody. Whole lanes were measured for the calculation of protein amounts using ImageJ. cFBPase detection was used for normalization. Error bars represent SD of n ≥ 2 biological replicates. Asterisk indicates significant difference compared to Col-0 (Student’s t test; *p<0.05, ***p<0.001).
-
Figure 3—source data 1
(D-E) Comparison of the amount of the total SUMOylation with and without cold treatment.
- https://doi.org/10.7554/eLife.44213.013
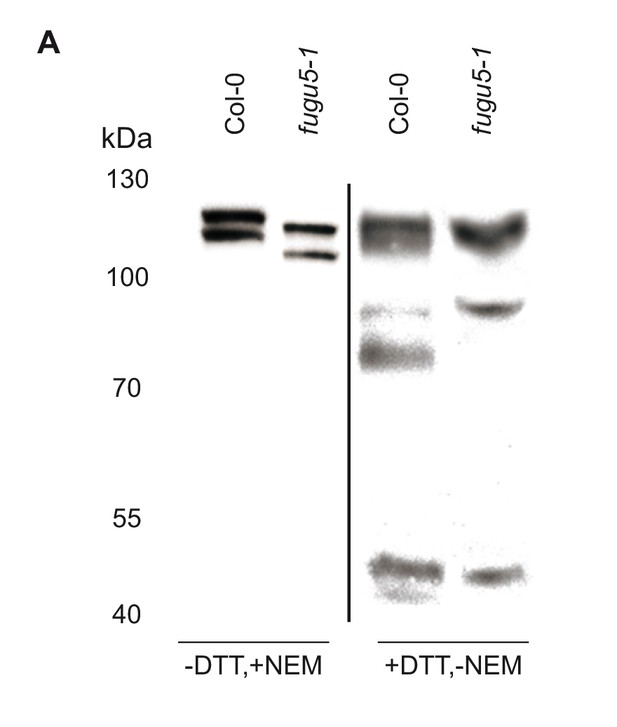
Western blot detection of ICE1 in protein extracted in the presence of DTT.
(A) Comparison of total protein of 10-days-old Col-0 and fugu5-1 seedlings extracted ±DTT (5 mM) and NEM (20 mM).
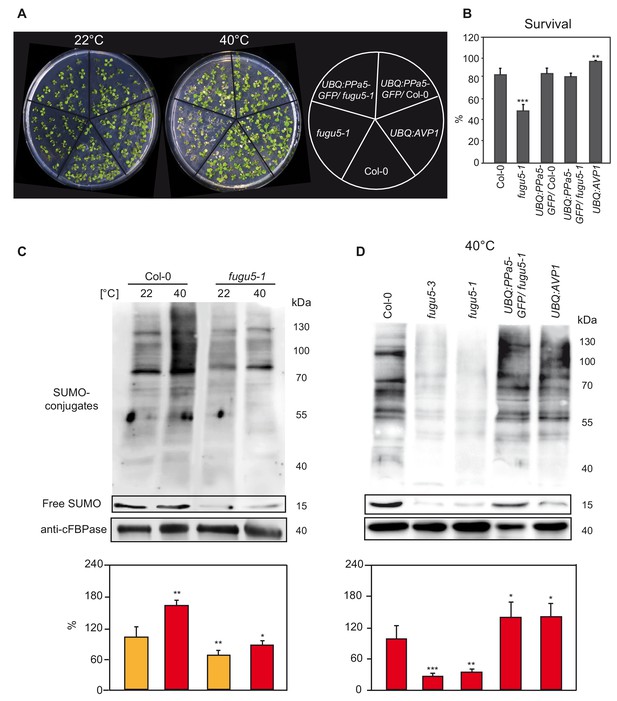
Heat shock-induced SUMOylation is also reduced in fugu5.
(A) Phenotypic analysis of 10-days-old Col-0, fugu5-1, UBQ:PPa5-GFP in Col-0 and fugu5-1 backgrounds and UBQ:AVP1 seedlings analysis before and after heat. Representative pictures of seedlings before and 4 days after completion of heat shock treatment (40°C, 4 hours) are depicted. (B) Seedling survival was determined 4 days after the heat shock. Alive and dead seedlings were counted and survival is shown as the percentage of the living seedlings. Error bars show SD of the mean with n ≥ 24 samples of one representative experiment. Two biological experiments were performed. Asterisk indicates significant difference compared to Col-0 (Student’s t test; **p<0.01, ***p<0.001). (C) SUMOylation levels of Col-0 and V-PPase mutant fugu5-1 were analysed with western blot under normal conditions (22°C) and after heat shock treatment (40°C, 30 min). (D) Measurement of the SUMO amount of Col-0, V-PPase mutants, UBQ:PPa5-GFP/fugu5-1 and UBQ:AVP1 seedlings after heat shock treatment (40°C, 30 min). (C) and (D) 10-days-old liquid grown seedlings are used for total protein extraction. Anti-SUMO1/2 (Agrisera) was used as primary antibody. Whole lanes were measured for the calculation of protein amounts using ImageJ. cFBPase detection was used for normalization. Error bars represent SD of n = 2 biological replicates. Asterisk indicates significant difference compared to Col-0 (Student’s t test; *p<0.05, **p<0.01, ***p<0.001).
-
Figure 4—source data 1
(B) Survival measurement of the 10 days old seedlings upon heat shock.
(C-D) Comparison of the amount of the total SUMOylation with and without heat shock treatment.
- https://doi.org/10.7554/eLife.44213.015
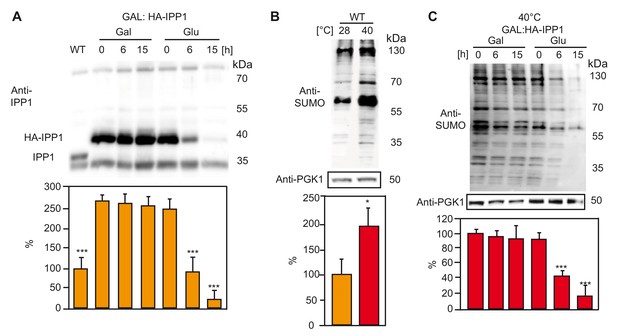
Increased PPi levels interfere with SUMOylation in yeast.
(A) Amount of the soluble pyrophosphatase protein in a conditional Ipp1 mutant of S. cerevisiae (GAL:HA-IPP1). Asterisk indicates significant difference compared to GAL:HA-IPP1 in galactose time point 0 hr (Student’s t test; ***p<0.001). Wt strain (W303) was used as a control. (B) Amount of total SUMOylation in W303 determined before and after heat stress. Asterisk indicates significant difference (Student’s t test; *p<0.05) (C) Measurement of total SUMO protein in conditional IPP1 mutant of S. cerevisiae (GAL:HA-IPP1). Asterisk indicates significant difference (Student’s t test; ***p<0.001). (A–C) Yeast is grown in synthetic complete medium supplemented with galactose at 28°C. After growing until OD600 0.5, part of the Ipp1 conditional mutant is switched to glucose supplemented medium to suppress the promoter and samples are collected at the indicated time points. For the heat treatment, cultures were switched to 40°C incubator for 1 hr. Error bars represent SD of n ≥ 2 biological replicates.
-
Figure 5—source data 1
(A) Amount of IPP1 in different carbon supplies over time.
(B) Comparison of the total SUMOylation of wt yeast at 28 and 40°C. (C) Comparison of the total SUMOylation at 40°C, in Ipp1 conditional mutant.
- https://doi.org/10.7554/eLife.44213.017
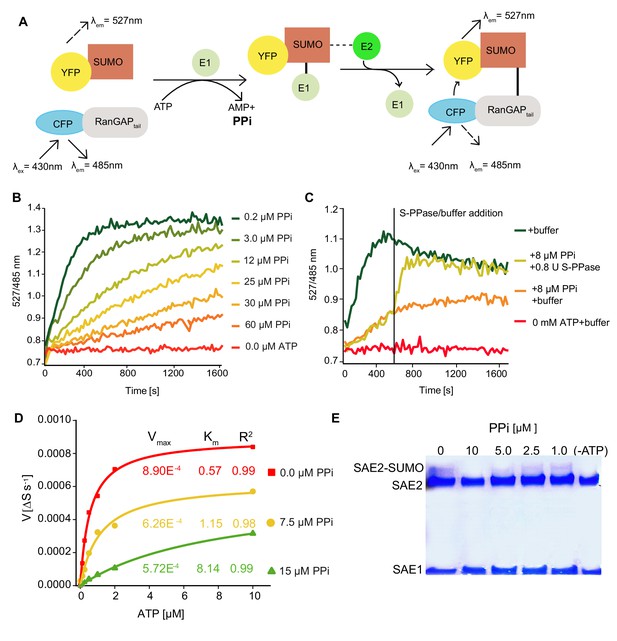
PPi regulates SUMOylation activity in vitro.
(A) Schematic illustration of the FRET-based sumoylation assays. Upon addition of ATP, the human SUMO E1 activating enzyme Aos1/Uba2 and the E2 conjugating enzyme Ubc9 form an isopeptide bond between the CFP-tagged human model substrate GAPtail and YFP-tagged mature SUMO. This can be detected via FRET measurements: Following the excitation of CFP, energy is transferred onto YFP. YFP and CFP emission are recorded upon excitation at 430 nm. Measurements are calculated as the ratio of the λem (YFP-SUMO 527 nm) to λex (CFP-RanGAPtail, 485 nm). (B) PPi titration showing that the increasing PPi concentration inhibits the SUMOylation activity. 1 mM ATP used for all the measurements. (C) In vitro SUMOylation assay showing that the E. coli soluble PPase is able to remove the inhibitory effect of PPi. After 10 min of measurement, one of the 8 µM PPi containing wells were supplied with 0.8 U of E.coli soluble PPase and control buffer was added to the rest of the wells. Measurements were continued for 20 more minutes. (D) PPi addition results in mixed inhibition of SUMOylation activity. Michaelis-Menten fittings of the measurements shown in Figure 6—figure supplement 1. Fittings are done in Origin software and Vmax and Km are calculated accordingly. (E) In vitro thioester bond formation assay showing Arabidopsis E1 (SAE2/SAE1a) and SUMO conjugation under different PPi concentrations. (B–D) Experiments were repeated four times, one representative experiment is shown.
-
Figure 6—source data 1
(B) Ratio values (527/485 nm) of the FRET based SUMOylation showing the effect of the increasing amount of PPi concentrations.
(C) Ratio values (527/485 nm) of the FRET based SUMOylation assay showing that theE. colisoluble PPase is able to remove the inhibitory effect of PPi. (D) Calculation of the Vmax and Km of SUMOylation activity upon PPi inhibition based on the data shown in Figure 6—figure supplement 1.
- https://doi.org/10.7554/eLife.44213.023
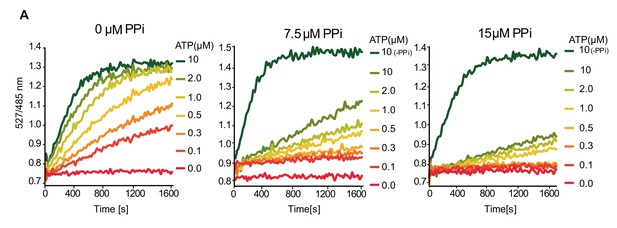
Elevated PPi concentrations leads to mixed inhibition of SUMOylation in vitro.
(A) SUMOylation assays demonstrating the effect of the increasing amounts of PPi to the speed of the reaction in a range of 0–10 µM ATP concentration. Experiments were repeated four times, one representative experiment is presented.
-
Figure 6—figure supplement 1—source data 1
Ratio values (527/485 nm) of the FRET based SUMOylation assay showing the effect of the increasing amounts of PPi to the speed of the reaction in a range of 0–10µMATP concentration.
- https://doi.org/10.7554/eLife.44213.020
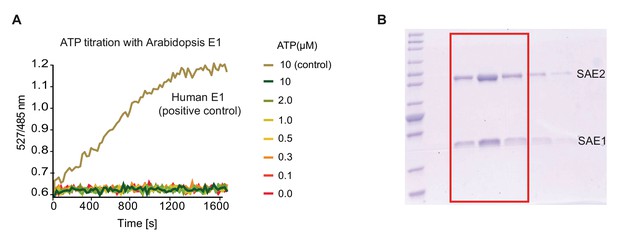
AtSAE1/2 purification and its activity in in vitro FRET based SUMOylation assay.
(A) Gel picture after purification of AtSAE1 and AtSAE2. Highlighted fractions from the final gel filtration step were combined, dialysed and used for subsequent experiments. (B) The purified Arabidopsis E1 activating enzyme is not functional in the FRET based SUMOylation assay. Assays were set up as described for Figure 6, but with recombinant Arabidopsis E1 enzyme. Human E1 activating enzyme was used in a positive control.
-
Figure 6—figure supplement 2—source data 1
(A) Ratio values (527/485 nm) of the FRET-based SUMOylation assay with Arabidopsis E1.
- https://doi.org/10.7554/eLife.44213.022
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Arabidopsis thaliana) | AtAVP1 | TAIR: AT1G15690 | ||
| Gene (Arabidopsis thaliana) | AtPPa1 | TAIR: AT1G01050 | ||
| Gene (Arabidopsis thaliana) | AtPPa2 | TAIR: AT2G18230 | ||
| Gene (Arabidopsis thaliana) | AtPPa4 | TAIR: AT3G53620 | ||
| Gene (Arabidopsis thaliana) | AtPPa5 | TAIR: AT4G01480 | ||
| Gene (Arabidopsis thaliana) | AtSUMO1 | TAIR: AT4G26840 | ||
| Gene (Arabidopsis thaliana) | AtICE1 | TAIR: AT3G26744 | ||
| Gene (Arabidopsis thaliana) | AtSAE1 | TAIR: AT4G24940 | ||
| Gene (Arabidopsis thaliana) | AtSAE2 | TAIR: AT2G21470 | ||
| Gene (Saccharomices cerevisiae) | ScIPP1 | SGD: YBR011C | ||
| Strain, strain background (Agrobacterium tumefaciens) | ASE | Lampropoulos et al., 2013 | pSOUP+ | |
| Strain, strain background (Saccharomices cerevisiae) | W303 | Szoradi et al., 2018 | SSY122 | |
| Strain, strain background (Saccharomices cerevisiae) | IPP1prΔ::HIS3- GAL1pr-HA-IPP1 | this paper | SSY2542 | Sebastian Schuck lab |
| Genetic reagent (Arabidopsis thaliana) | fugu5-1 | Ferjani et al., 2011 | ||
| Genetic reagent (Arabidopsis thaliana) | fugu5-3 | Ferjani et al., 2011 | ||
| Genetic reagent (Arabidopsis thaliana) | ice1-2 | Nottingham Arabidopsis Stock Centre (NASC) | SALK_003155 | |
| Genetic reagent (Arabidopsis thaliana) | ppa1 | Segami et al., 2018; NASC | SAIL_251_D07 | |
| Genetic reagent (Arabidopsis thaliana) | ppa1,4 | this paper | SAIL_251_D07, SAIL_916_C08 | Cross between the mutants ppa1 and ppa4 described in Segami et al., 2018 Masayoshi Maeshima lab |
| Genetic reagent (Arabidopsis thaliana) | ppa1,2,4,5 | Segami et al., 2018 | SAIL_251_D07, SAIL_618_H05, SAIL_916_C08, SALK_014647 | |
| Biological sample (Arabidopsis thaliana) | AVP1:IPP1/fugu5-1 | Ferjani et al., 2011 | AVP1 promoter:IPP1 coding sequence; fugu5-1 mutant background | |
| Biological sample (Arabidopsis thaliana) | UBQ:AVP1 #18–4 | Kriegel et al., 2015 | UBQ promoter:AVP1 coding sequence; Col-0 wild-type background | |
| Biological sample (Arabidopsis thaliana) | UBQ:PPa5-GFP/Col-0 | this paper, | UBQ promoter:PPa5-GFP coding sequence; Col-0 wild-type background. Karin Schumacher lab | |
| Biological sample (Arabidopsis thaliana) | UBQ:PPa5-GFP/fugu5-1 | this paper, | UBQ promoter:PPa5-GFP coding sequence; fugu5-1 wild-type background. Karin Schumacher lab | |
| Antibody | Anti-SUMO1 (rabbit polyclonal) | Agrisera | AS08308 | (1:1000) |
| Antibody | Anti-ICE1 (rabbit polyclonal) | Agrisera | AS163971 | (1:1000) |
| Antibody | Anti-cFBPase (rabbit polyclonal) | Agrisera | AS04043 | (1:5000) |
| Antibody | Anti-rabbit-HRP (Goat polyclonal) | Promega | W401B | (1:10000) |
| Antibody | Anti-IPP1 (rabbit polyclonal) | Antibodies- online GmbH | ABIN459215 | (1:1000) |
| Antibody | Anti-PGK1 (mouse monoclonal) | Abcam | AB113687 | (1:100000) |
| Antibody | Anti-mouse-HRP (sheep polyclonal) | GE Healthcare UK | NXA931 | (1:5000) |
| Antibody | Anti-V-PPase (rabbit polyclonal) | Agrisera | AS121849 | (1:10000) |
| Antibody | Anti-VHA-C (rabbit polyclonal) | Karin Schumacher lab | Schumacher et al., 1999 | (1:2000) |
| Antibody | Anti-GFP (rabbit polyclonal) | Karin Schumacher lab | Roth et al., 2018 | (1:10000) |
| Recombinant DNA reagent | UBQ:PPa5-GFP | this paper, | vector-promoter:tagged-protein construct. Karin Schumacher lab | |
| Recombinant DNA reagent | pET28b(+)−6xHIS -AtSUMO1(1–93) | this paper, | vector-tagged-protein construct. Karin Schumacher lab | |
| Recombinant DNA reagent | pET28-6xHIS-AtSAE1 | this paper, | vector-tagged-protein construct. Frauke Melchior lab | |
| Recombinant DNA reagent | pET11d-AtSAE2 | this paper | vector-protein construct. Frauke Melchior lab | |
| Recombinant DNA reagent | pFA6a-His3M × 6- PGAL1 | this paper, | vector-promoter:tagged-protein construct. Sebastian Schuck lab | |
| Peptide, recombinant protein | CFP-RanGAPtail | Bossis et al., 2005; Werner et al., 2009 | ||
| Peptide, recombinant protein | YFP-SUMO | Bossis et al., 2005; Werner et al., 2009 | ||
| Peptide, recombinant protein | Uba2/Aos1 | Bossis et al., 2005; Werner et al., 2009 | ||
| Peptide, recombinant protein | Ubc9 | Bossis et al., 2005; Werner et al., 2009 | ||
| Peptide, recombinant protein | 6xHis-AtSUMO1 | this paper | For in vitro thioester bond formation assays | |
| Peptide, recombinant protein | AtSAE1 | this paper | For in vitro thioester bond formation assays | |
| Peptide, recombinant protein | AtSAE2 | this paper | For in vitro thioester bond formation assays | |
| Commercial assay or kit | SPL kit | NH DyeAgnostics | Western blot protein quantification kit |
Additional files
-
Supplementary file 1
Additional resources.
(A) Primers of UBQ:PPa5-GFP construct. (B) List of GG modules. (C) qRT primers. (D) Primers for constructs used in protein purification. (E) Statistical analysis (One-way ANOVA followed by Tukey’s test, p<0.05) of the electrolyte leakage assay (Figure 1C). Significant values are highlighted.
- https://doi.org/10.7554/eLife.44213.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44213.025





