Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism
Figures
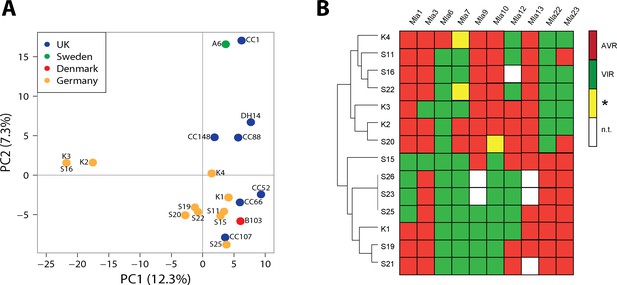
Population structure and avirulence profiles of Blumeria graminis f. sp. hordei (Bgh) isolates.
(A) Principal Component Analysis (PCA) of the indicated European Bgh isolates, including ten newly collected strains from a local pathogen population in Germany, based on a set of 5170 diallelic high-quality synonymous SNPs. (B) Hierarchical clustering (R package 'pheatmap') of avirulence profiles from 14 Bgh isolates collected within the same area near Cologne, Germany (GPS 5˚57’N, 6˚51’E 5). Numbers correspond to infection types (ITs) 1, 2 and 3 = avirulent, red; infection types 4 and 5 = virulent, green. *denotes differences of ITs between cultivars Pallas and Manchuria, yellow. n.t.: not tested, white.
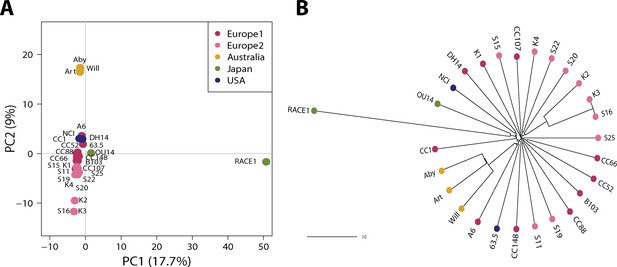
Population structure of a worldwide collection of 27 Blumeria graminis f. sp. hordei (Bgh) isolates.
Principal Component (PC)Analysis (A) and Neighbour-joining (B) tree of a worldwide collection of Bgh isolates including the ten newly collected Bgh isolates generated on the basis of a set of 6286 diallelic high-quality synonymous SNPs.
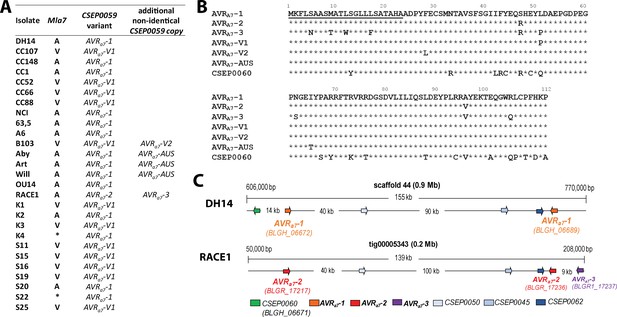
Identification of CSEP0059 as an AVRa7 candidate by association of avirulence profiles with transcript polymorphisms and integration in the physical Bgh map.
(A) AVRa7 transcript variants encoded by the indicated Bgh isolates with corresponding avirulence profiles. (B) Alignment of deduced AVRA7 amino acid sequences with all variants highlighted. (C) Visualization of the chromosomal regions harboring CSEP0059/AVRa7 candidate variants with corresponding gene IDs in the genomes of Bgh isolates DH14 and RACE1. All CSEPs are depicted by arrows. * denotes different infection types between cultivars Pallas and Manchuria.
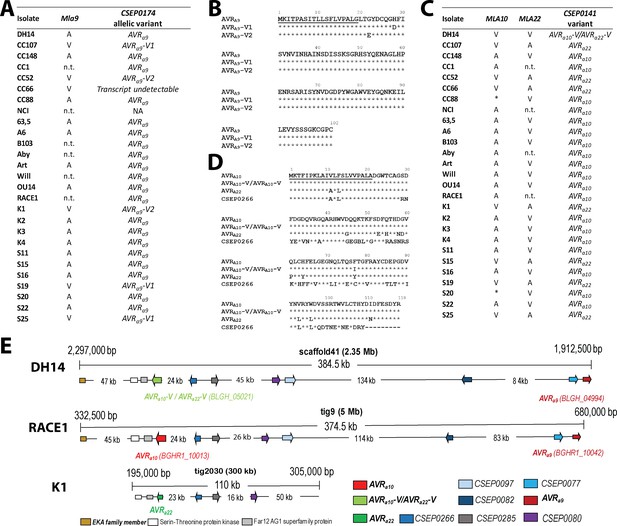
Identification of CSEP0174 as an AVRa9 candidate and CSEP0141 as an AVRa10 and AVRa22 candidate by association of avirulence profiles with transcript polymorphisms as well as integration in the physical Bgh map.
(A) AVRa9 variants encoded by each Bgh isolate with corresponding avirulence profiles. (B) Alignment of deduced AVRA9 amino acid sequences with variants highlighted. (C) AVRa10/AVRa22 variants encoded by each Bgh isolate with corresponding avirulence profiles. (D) Alignment of AVRA10, AVRA22, and AVRA10-V/AVRA22-V amino acid sequences. (E) Visualization of the chromosomal regions harboring CSEP0174/AVRa9 and CSEP0141/AVRa10/AVRa22 candidates with corresponding gene IDs as well as a copy of the EKA family class-1 retrotransposon and other CSEPs in the genomes of Bgh isolates DH14, RACE1, and K1. *denotes differences of infection types between cultivars Pallas and Manchuria. n.t.: not tested.
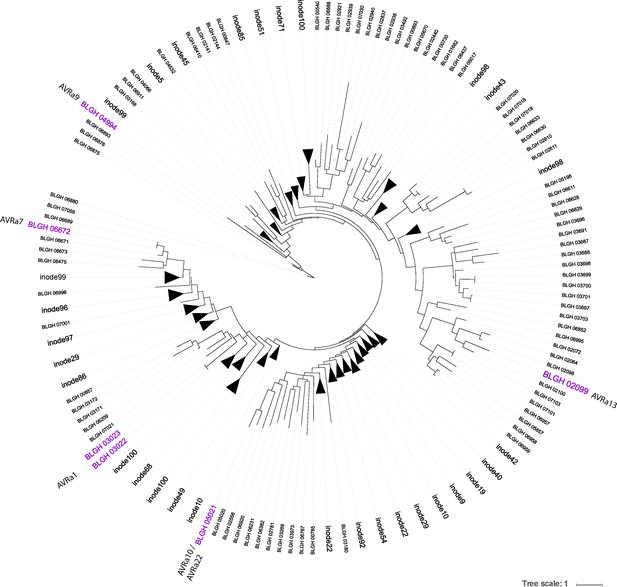
Maximum likelihood phylogeny tree for the 805 predicted secreted proteins of Bgh DH14 lacking respective signal peptides.
Isolated AVRas are highlighted in purple; triangles depicting nodes, which are collapsed to allow a more compact visualization of the tree are proportional to the number of edges they contain.
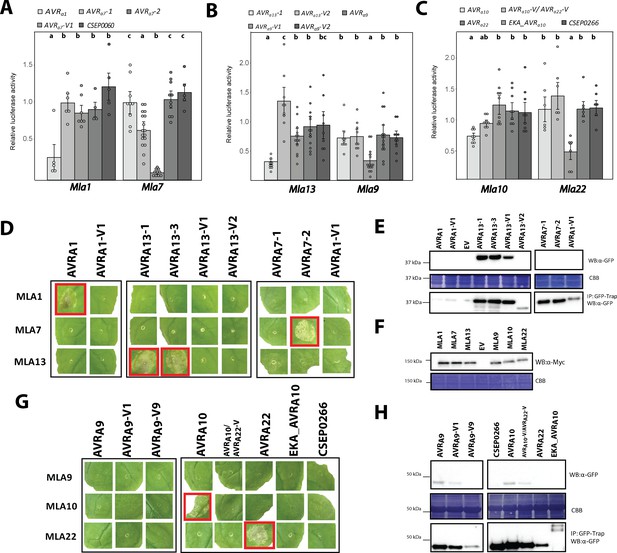
AVRA candidates induce MLA-specified cell death in transient gene expression assays.
(A-C) Barley protoplasts were transfected with pUBQ:luciferase and either an piPKb002 empty vector control (EV) or piPKb002 containing cDNAs of (A) AVRa1, AVRa7 variants and CSEP0060, all lacking their respective signal peptides (SPs) together with either Mla1 or Mla7; (B) AVRa9 variants and AVRa13 variants, all lacking their respective SPs together with either Mla9 or Mla13; (C) AVRa10, AVRa10-V/AVRa22-V, AVRa22, and CSEP0266 without SPs and EKA_AVRa10, together with either Mla10 or Mla22. Luciferase activity was determined ~16 hr post transfection as a proxy for cell death and normalized for each Mla construct by setting the detected luminescence for the corresponding EV transfection to 1. All values obtained in at least four independent experiments are indicated by dots, error bars = standard deviation. Differences between samples were assessed by analysis of variance (ANOVA) and subsequent Tukey post hoc tests for each Mla construct. Calculated P values were as follows: Mla7: p=1.2e-13 and Mla1: p=1.9e-03 (A); Mla13: p=3.2e-06 and Mla9: p=1.3e-04 (B); Mla10: p=4.2e-04 and Mla22: p=5.5e-04 (C). Samples marked by identical letters in the plots do not differ significantly (p<0.05) in the Tukey test for the corresponding transfected Mla. (D–H) Nicotiana benthamiana plants were transformed transiently as indicated. cDNAs lacking stop codons were fused in between the 35S promotor sequence and 4xMyc (Mla variants) or mYFP (CSEPs and AVRa variants lacking SPs and EKA_AVRa10) epitope sequences. Cell death (D, G) was determined three days post transformation and figures shown are representatives of at least three independently performed experiments with at least three transformations per experiment. Protein levels (E, F, H) of MLA-4xMyc, CSEP-mYFP, AVRA-mYFP and EKA_AVRA-mYFP in Nicotiana benthamiana corresponding to constructs of D and G. Leaf tissue was harvested two days post infiltration. Total protein was extracted and recovered by GFP-Trap pull-down as indicated. Extracts and immunoprecipitates were separated by gel electrophoresis and probed by anti-Myc or anti-GFP western blotting (WB) as indicated. IP: Immunoprecipitated fraction. CBB: Coomassie brilliant blue.
-
Figure 4—source data 1
Data points indicating relative luciferase activity of Figure 4A.
- https://doi.org/10.7554/eLife.44471.011
-
Figure 4—source data 2
Data points indicating relative luciferase activity of Figure 4B.
- https://doi.org/10.7554/eLife.44471.012
-
Figure 4—source data 3
Data points indicating relative luciferase activity of Figure 4C.
- https://doi.org/10.7554/eLife.44471.013
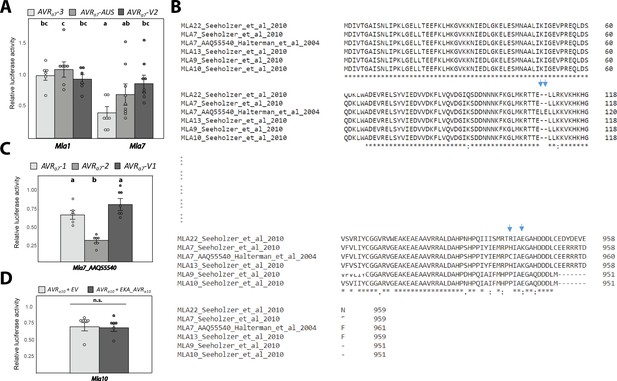
MLA-mediated cell death in barley protoplasts.
(A) Barley protoplasts were transfected with pUBQ:luciferase together with either an empty vector control (EV) or the AVRa7-3, AVRa7-V2, AVRa7-AUS constructs all lacking their respective signal peptides (SPs) together with either Mla1 or Mla7. (B) Amino acid sequence alignment of Uniprot MLA7_ AAQ55540 (Halterman and Wise, 2004) with MLA alleles obtained by translation of nucleotide sequences from respective Manchuria Mla amplicons (Seeholzer et al., 2010). Blue arrows indicate atypical amino acid substitutions and insertion of the Uniprot_AAQ55540/NCBI_AY266444 MLA7 protein sequence. (C) Barley protoplasts were transfected with pUBQ:luciferase together with either EV or cDNAs of AVRa7-1, AVRa7-2, or AVRa7-V1 all lacking their respective SPs together with Mla7_ AAQ55540 (NCBI_GeneBank: AY266444). (D) Barley protoplasts were transfected with pUBQ:luciferase together with cDNAs of Mla10 and AVRa10 lacking their respective signal peptides (4 µg) and either 4 µg of EV or 4 µg of EKA_AVRa10 constructs. Luciferase activity (A, C, D) was determined 16 hr post transfection as a proxy for cell death and normalized for each Mla construct by setting the detected luminescence for the corresponding empty vector transfection to 1. n ≥ 6, error bars = standard deviation. Differences between relative luciferase activities of individual samples were assessed by using analysis of variance (ANOVA). Calculated P values were as follows: (A) p=8.6e-05; (B) p=2.5e-06; (C) p=0.81 (D). Samples marked by identical letters in the plot did not differ significantly (p<0.05) in the Tukey test. n.s. = not significant.
-
Figure 4—figure supplement 1—source data 1
Data points indicating relative luciferase activity of Figure 4—figure supplement 1A.
- https://doi.org/10.7554/eLife.44471.014
-
Figure 4—figure supplement 1—source data 2
Data points indicating relative luciferase activity of Figure 4—figure supplement 1C.
- https://doi.org/10.7554/eLife.44471.015
-
Figure 4—figure supplement 1—source data 3
Data points indicating relative luciferase activity of Figure 4—figure supplement 1D.
- https://doi.org/10.7554/eLife.44471.016
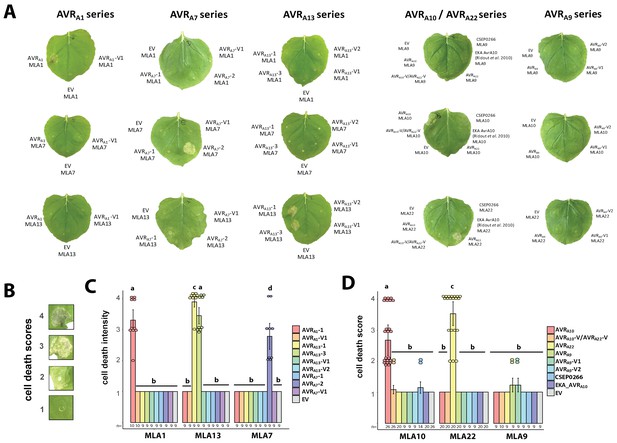
AVRA-mYFP candidates induce MLA-4xMyc mediated cell death.
(A) Leaves of transiently transformed Nicotiana benthamiana plants corresponding to Figure 4D and G. ( B–D) Scoring of cell death from 1 (no cell death) to 4 (cell death of full infiltration zone) (B) of N. benthamiana leaves transiently transformed with constructs encoding Mla1-4xMyc, Mla7-4xMyc, or Mla13-4xMyc together with AVRa1-1, AVRa1-V1, AVRa13-1, AVRa13-3, AVRa13-V1, AVRa13-V2, AVRa7-1, AVRa7-2, or AVRa7-V1 constructs all lacking signal peptides (SPs) and fused C-terminally in frame with a mYFP tag (C), and Mla9-4xMyc, Mla10-4xMyc, or Mla22-4xMyc together with AVRa9, AVRa9-V1, AVRa9-V2, AVRa10, AVRa10-V/AVRa22-V, AVRa22, or CSEP0266 constructs without SP or EKA_AVRa10, all fused C-terminally to a mYFP tag (D). Pictures for cell death scoring were taken three days post transformation. Data were obtained from at least three independently performed experiments with at least three transformations per experiment (n as indicated and ≥9). Dots = scores beyond 1, error bars = standard deviation. Differences between cell death scores of samples were assessed by using analysis of variance (ANOVA) and subsequently Tukey post hoc tests. Calculated P values: P =<2.2e-16 (C and D). Samples marked by identical letters do not differ significantly (p>0.05) in the Tukey test.
-
Figure 4—figure supplement 2—source data 4
Ceall death scores of samples shown in Figure 4D according to scoring system of Figure 4—figure supplement 2B.
- https://doi.org/10.7554/eLife.44471.017
-
Figure 4—figure supplement 2—source data 5
Ceall death scores of samples shown in Figure 4G according to scoring system of Figure 4—figure supplement 2B.
- https://doi.org/10.7554/eLife.44471.018
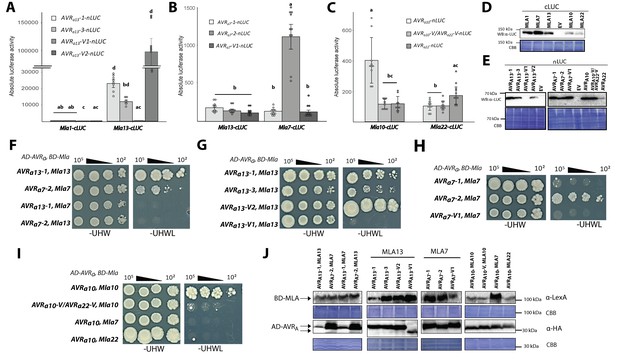
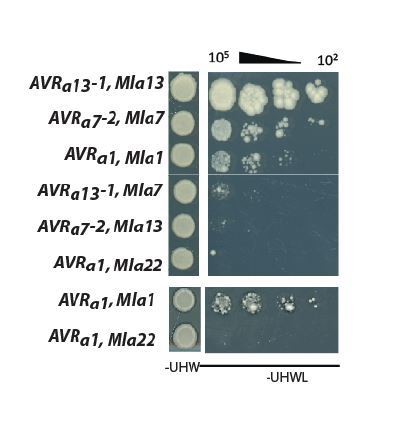
Association of candidate AVRA proteins with MLA in plant extracts (A–E) and in yeast (F–J).
(A-C) Nicotiana benthamiana plants were transformed transiently with constructs encoding (A) Mla1-cLUC or Mla13-cLUC together with cDNAs of AVRa13-1, AVRa13-3, AVRa13-V1, AVRa13-V2 lacking signal peptides (SPs) and fused C-terminally in frame with nLUC, (B) Mla1-cLUC, or Mla7-cLUC together with cDNAs of AVRa7-1, AVRa7-2, and AVRa7-V1 lacking SPs and fused C-terminally in frame with nLUC, (C) Mla10-cLUC or Mla22-cLUC together with cDNAs of AVRa10, AVRa10-V/AVRa22-V, and AVRa22 without SPs fused C-terminally in frame with nLUC, all under the control of the 35S promotor. Luciferase activity was determined two days post transfection. All values obtained in at least six experiments are indicated by dots, error bars = standard deviation. For each graph, differences between samples were assessed using non-paramatric analysis of variance (Kruskal-Wallis) and subsequent Dunn’s post hoc tests. Calculated P values were as follows: (A) p=6.8e-10, (B) p=1.2e-04, (C) p=8.0e-07. Samples marked by identical letters in the plot did not differ significantly (p<0.05) in Dunn’s test. (D–E) Protein levels of MLA-cLUC (D) and AVRA-nLUC (E) variants in Nicotiana benthamiana corresponding to constructs of 5A to 5C. Leaf tissue was harvested two days post infiltration. Total protein was extracted, separated by gel electrophoresis and probed by anti-LUC western blotting (WB). (E–I) Yeast cells were co-transformed with Mla alleles fused N-terminally to the LexA binding domain (BD) and AVRa constructs lacking SPs fused N-terminally to the B42 activation domain (AD) and 1xHA tag as indicated. Growth of transformants was determined on selective growth media containing raffinose and galactose as carbon sources but lacking uracil, histidine and tryptophan (-UHW), and interaction of proteins was determined by leucine reporter activity reflected by growth of yeast on selective media containing raffinose and galactose as carbon sources but lacking uracil, histidine, tryptophan and leucine (-UHWL). Figures shown are representatives of at least three independent experiments with yeast clones obtained from three independent yeast transformation experiments and pictures were taken 12 days after drop out. (J) Protein levels of BD-MLA and AD-AVRA variants corresponding to yeast of Figure 5D–5G. Yeast transformants were grown in glucose containing selective media lacking uracil, tryptophan, and histidine to OD600 = 1. Cells were harvested, total protein extracted, separated by gel electrophoresis, and western blots (WB) were probed with anti-LexA or anti-HA antibodies as indicated.
-
Figure 5—source data 1
Data points indicating absolute luciferase activity of Figure 5A.
- https://doi.org/10.7554/eLife.44471.020
-
Figure 5—source data 2
Data points indicating absolute luciferase activity of Figure 5B.
- https://doi.org/10.7554/eLife.44471.021
-
Figure 5—source data 3
Data points indicating absolute luciferase activity of Figure 5C.
- https://doi.org/10.7554/eLife.44471.022
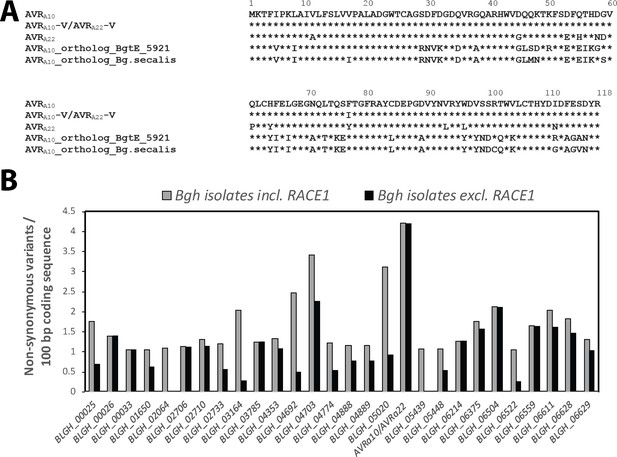
Conservation of AVRa10/AVRa22 orthologs between Blumeria graminis formae speciales.
(A) Alignment of protein sequences (AVRA10/AVRA22) encoded by Bgh CSEP0141 and orthologs detected in B. graminis f. sp. tritici and B. graminis f. sp. secalis. (B) Number of non-synonymous sequence variants was determined for 190 core effectors (Frantzeskakis et al., 2018) among all Bgh isolates described in this study and is displayed for all core effectors with ≥1 non-synonymous variants/100 bp coding sequence. Grey bars, including all Bgh isolates; black bars, all Bgh isolates excluding RACE1.
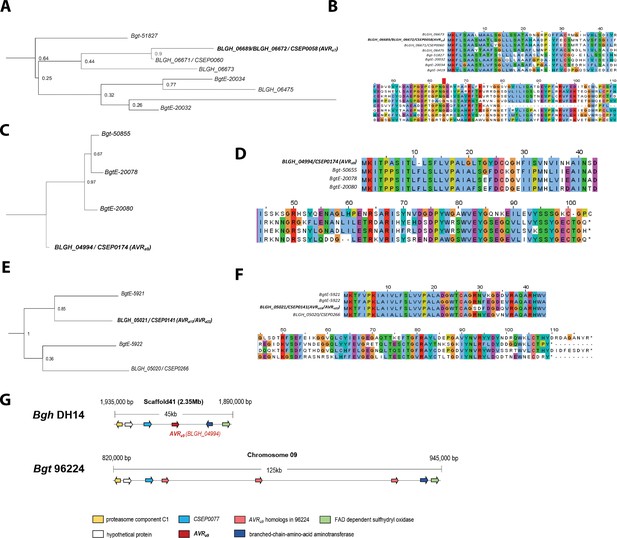
Phylogenetic and genomic analysis of AVRa7, AVRa9, AVRa10/AVRa22 homologs in Blumeria graminis f. sp. tritici.
(A-F) Maximum-likelihood trees (A, C, E) with labelled bootstrap percentages and corresponding sequence alignments (B, D, F) based on deduced protein sequences encoded by (A, B) CSEP0059 (AVRa7) (C, D) CSEP0174 (AVRa9) and (E, F) CSEP0141 (AVRa10/AVRaa22). (G) Visualization of the genomic region harboring CSEP0174/AVRaa9 in the genomes of B. graminis f. sp. hordei isolate DH14 and B. graminis f. sp. tritici isolate 96224.
Tables
AVRa10/AVRa22 ortholog BgtE-5921 variants in different Blumeria graminis formae speciales.
https://doi.org/10.7554/eLife.44471.025| Number of variants in Blumeria graminis f. sp. | |||||
|---|---|---|---|---|---|
| AVRa10/AVRa22 ortholog BgtE-5921 | tritici | dicocci/tritici2 | triticale | secalis | Total |
| Hap_96224 | 29 | 22 | 51 | ||
| Hap_96224 R111G | 2 | 2 | |||
| Hap_96224 V5I, R111G | 2 | 2 | |||
| Hap_96224 R111G, A115G | 1 | 1 | |||
| Hap_96224 F77L, R111G | 4 | 4 | |||
| Hap_96224 F77L, R111G, Y117H | 1 | 1 | |||
| Hap_96224 V5I, I8L, F77L, R111G | 1 | 1 | |||
| Hap_96224 V17I, S47M, D48N, R50K, G58D, G59S, R101C, R111G, A115V | 5 | 5 | |||
| total number of isolates | 34 | 6 | 22 | 5 | 67 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain (Blumeria graminis f. sp. hordei) | CC107, CC148, CC1, CC52, CC66, CC88, NCI, 63.5, A6, B103, Aby, Art, Will, OU14, RACE1, K1 | Lu et al. (2016) doi:10.1073/pnas.1612947113. | GEO:GSE83237 | |
| Strain (Blumeria graminis f. sp. hordei) | DH14 | Frantzeskakis et al. (2018) doi:10.1186/s12864-018-4750-6. | GEO:GSE106282 | |
| Strain (Blumeria graminis f. sp. hordei) | K2, K3, K4, S11, S15, S16, S19, S20, S21, S22, S23. S25, S26 | this paper | GEO:GSE110266 | collected in 2017 on cv. Meridian and Keeper barley at the Max Planck Institute for Plant Breeding Research, Cologne, Germany (GPS 5˚57′N, 6˚51′E 5) |
| Recombinant DNA reagent | pIPKb002 | Himmelbach et al. (2007) doi:10.1104/pp.107.111575. | NCBI:EU161568.1 | pZmUBQ:GW, SpcR |
| Recombinant DNA reagent | pGWB517 | Nakagawa et al. (2007) doi:10.1263/jbb.104.34. | NCBI:AB294484.1 | p35S:GW-4Myc, SpcR |
| Recombinant DNA reagent | pXCSG-GW-mYFP | García et al. (2010) doi:10.1371/journal.ppat.1000970. | NA | p35S:GW-mYFP, CarbR |
| Recombinant DNA reagent | pB42AD-GW | Shen et al. (2007) doi:10.1126/science.1136372. | NA | pGal1:B42-AD-−1xHA-GW, TRP |
| Recombinant DNA reagent | pLexA-GW | Shen et al. (2007) doi:10.1126/science.1136372. | NA | pADH1:LexA-BD-GW, HIS3 |
| Recombinant DNA reagent | pDest-GW-nLUC | Gehl et al. (2011) doi:10.1111/j.1365-313X.2011.04607.x. | NA | p35S:GW-NterminusLuciferase, KanR |
| Recombinant DNA reagent | pDest-GW-cLUC | Gehl et al. (2011) doi:10.1111/j.1365-313X.2011.04607.x. | NA | p35S:GW-CterminusLuciferase, KanR |
| Gene (Blumeria graminis f. sp. hordei) | AVRa7 variants | Frantzeskakis et al. (2018) doi:10.1186/s12864-018-4750-6. | csep0059; BLGH_06689; BLGH_06672; BGHR1_17217; BGHR1_17236; BGHR1_17237 | |
| Gene (Blumeria graminis f. sp. hordei) | AVRa9 variants | Frantzeskakis et al. (2018) doi:10.1186/s12864-018-4750-6. | csep0174; BLGH_04994; BGHR1_10042 | |
| Gene (Blumeria graminis f. sp. hordei) | AVRa10/AVRaa22 variants | Frantzeskakis et al. (2018) doi:10.1186/s12864-018-4750-6 | csep0141; BLGH_05021; BGHR1_10013 | |
| Gene (Blumeria graminis f. sp. hordei) | AVRa1 variants | Lu et al. (2016) doi:10.1073/pnas.1612947113; Frantzeskakis et al. (2018) doi:10.1186/s12864-018-4750-6 | csep0008; BLGH_03023; BLGH_03022; BGHR1_11142 | |
| Gene (Blumeria graminis f. sp. hordei) | AVRa13 variants | Lu et al. (2016) doi:10.1073/pnas.1612947113; Frantzeskakis et al. (2018) doi:10.1186/s12864-018-4750-6 | csep0372; BLGH_02099; BGHR1_12484 | |
| Gene (Hordeum vulgare) | Mla9 | Seeholzer et al. (2010) doi.org/10.1094/MPMI-23-4-0497 | NCBI: GU245941.1 | |
| Gene (Hordeum vulgare) | Mla22 | Seeholzer et al. (2010) doi.org/10.1094/MPMI-23-4-0497 | NCBI:GU245946 | |
| Gene (Hordeum vulgare) | Mla10 | Seeholzer et al. (2010) doi.org/10.1094/MPMI-23-4-0497 | Mla10 | Different from NCBI:AY266445.1 |
| Gene (Hordeum vulgare) | Mla7 | Seeholzer et al. (2010) doi.org/10.1094/MPMI-23-4-0497; Lu et al. (2016) doi:10.1186/s12864-018-4750-6 | Mla7 | Different from NCBI:AY266444.1 |
| Gene (Hordeum vulgare) | Mla7 (AAQ55540_Halterman et al., 2004) | Halterman and Wise (2004) doi:10.1111/j.1365-313X.2004.02032.x | NCBI:AY266444.1 | |
| Gene (Hordeum vulgare) | Mla1 | Seeholzer et al. (2010) doi.org/10.1094/MPMI-23-4-0497; Lu et al. (2016) | NCBI:GU245961 | |
| Gene (Hordeum vulgare) | Mla13 | Seeholzer et al. (2010), Lu et al. (2016) doi:10.1073/pnas.1612947113. | AF523678.1 | |
| Antibody | monoclonal rat anti-HA | Merck | 3F10, RRID:AB_390914 | 1:2000 |
| Antibody | monoclonal mouse anti-LexA | Santa Cruz Biotechnology | sc7544, RRID:AB_627883 | 1:1000 |
| Antibody | polyclonal rabbit anti-c-myc | Abcam | ab9106, RRID:AB_307014 | 1:5000 |
| Antibody | polyclonal rabbit anti-GFP | Abcam | ab6556, RRID:AB_305564 | 1:5000 |
| Antibody | polyclonal rabbit anti-LUC | Sigma | L0159, RRID:AB_260379 | 1:2000 |
| Antibody | polyclonal goat anti-rat IgG-HRP | Santa Cruz Biotechnology | sc2065, RRID:AB_631756 | 1:100 000 |
| Antibody | polyclonal goat anti-mouse IgG-HRP | Santa Cruz Biotechnology | sc2005, RRID:AB_631736 | 1:100 000 |
| Antibody | polyclonal donkey anti-rabbit IgG-HRP | Santa Cruz Biotechnology | sc-2313, RRID:AB_641181 | 1:100 000 |
| Antibody | monoclonal rabbit anti-GFP | Santa Cruz Biotechnology | sc-8334, RRID:AB_641123 | 1:5000 |
Additional files
-
Supplementary file 1
Infection phenotypes of Bgh isolates on the Pallas and Manchuria cultivar accessions used for the association test.
- https://doi.org/10.7554/eLife.44471.026
-
Supplementary file 2
Script for freebayes genetic association analysis.
- https://doi.org/10.7554/eLife.44471.027
-
Supplementary file 3
Script for mpileup genetic association analysis.
- https://doi.org/10.7554/eLife.44471.028
-
Supplementary file 4
Statistical summary of association analysis.
- https://doi.org/10.7554/eLife.44471.029
-
Supplementary file 5
Blumeria graminis isolates used for phylogenetic analysis of CSEP0141 (AVRa10/AVRaa22).
- https://doi.org/10.7554/eLife.44471.030
-
Supplementary file 6
Primers used in this study.
- https://doi.org/10.7554/eLife.44471.031
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44471.032