Optical estimation of absolute membrane potential using fluorescence lifetime imaging
Figures
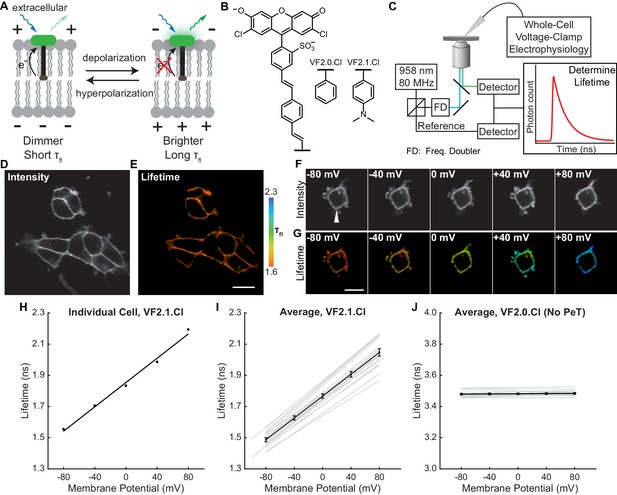
VoltageFluor FLIM linearly reports absolute membrane potential.
(A) Mechanism of VoltageFluor dyes, in which depolarization of the membrane potential attenuates the rate of photoinduced electron transfer. (B) Structures of the VF molecules used in this study. (C) Schematic of the TCSPC system used to measure fluorescence lifetime. Simultaneous electrophysiology was used to establish lifetime-voltage relationships. (D) Fluorescence intensity and (E) lifetime of HEK293T cells loaded with 100 nM VF2.1.Cl. (F) Intensity and (G) lifetime images of HEK293T cells voltage clamped at the indicated membrane potential. (H) Quantification of the single trial shown in (G), with a linear fit to the data. (I) Evaluation of VF2.1.Cl lifetime-voltage relationships in many individual HEK293T cells. Gray lines represent linear fits on individual cells. Black line is the average lifetime-voltage relationship across all cells (n = 17). (J) VF2.0.Cl lifetime does not exhibit voltage-dependent changes. Gray lines represent linear fits on individual cells, and the black line is the average lifetime-voltage relationship across all cells (n = 17). Scale bars represent 20 μm. Error bars represent mean ± SEM.
-
Figure 1—source data 1
Comparison of available approaches for measuring membrane potential in cells.
aMeasurements vary too much to be converted to absolute voltage or interpreted across populations of cells. This variability is attributable to numerous confounding factors, including dye loading, photobleaching, and sample movement (Peterka et al., 2011). bWhile in principle less variable than a single-color fluorescence intensity measurement, in practice, the signal depends strongly on the loading of two independent lipophilic indicators (Adams and Levin, 2012; Maher et al., 2007), which can vary substantially. cANEPPS excitation ratios depend on a variety of non-voltage factors, in particular the membrane composition, leading to substantial artifacts in optical Vmem determinations (Zhang et al., 1998; Gross et al., 1994). dWith the GEVI CAESR in our hands, apparently poor protein trafficking produces large amounts of non-voltage-sensitive signal, which contaminates the FLIM recording and contributes to high cell to cell variability (Figure 1—figure supplement 4, Materials and methods). ePatch-clamp electrophysiology requires physical contact with the cell of interest, which causes damage to the cell and, in whole cell configurations, washout of intracellular factors. Slight movement of the cell or sample generally result in loss of the patch. fMovement of the cell and photobleaching of the dye both cause large changes to the signal over seconds to minutes. gRatio-calibrated imaging approaches use a second signal (usually another color of fluorescence) to correct for differences in dye concentration or changes in the region of interest that contaminate single-color intensity signals. If the rate of photobleaching is the same for both components, photobleaching artifacts can also be avoided. hLimited by photon count rates. iLimited by probe movement in the membrane, which depends mostly on lipophilicity (Briggman et al., 2010). jPhoton counting based lifetime imaging, like epifluorescence intensity imaging, is limited by photon count rates. Large numbers of photons per pixel must be collected to fit TCSPC FLIM data, often using a line scanning confocal approach, leading to slower acquisition speeds than epifluorescence-based intensity imaging. kToxicity from capacitive load of the sensor (Briggman et al., 2010). lThe spatial resolution of electrophysiology is compromised by space clamp error, preventing interpretation of Vmem in regions far from the electrode (e.g. many neuronal processes) (Williams and Mitchell, 2008; 35,36). mAs demonstrated by Cohen and co-workers (Brinks et al., 2015); in our hands with CAESR, we also experienced significant improvements in voltage resolution by fitting a single curve per FLIM image instead of processing the images pixel-wise (see Materials and methods) nIn this work, we calibrated VF-FLIM for Vmem measurements with single cell resolution. In principle, subcellular spatial resolution could be achieved with the VF-FLIM technique.
- https://doi.org/10.7554/eLife.44522.008
-
Figure 1—source data 2
Properties of lifetime standards and VoltageFluor dyes.
Fluorescein and erythrosin B standards were measured in drops of solution placed on a coverslip. For VF dyes, voltage sensitivities from intensity-based fluorescence imaging in HEK293T cells (%ΔF/F, percent change in fluorescence intensity for a voltage step from −60 mV to +40 mV) are from previously published work (Woodford et al., 2015). Lifetime data were obtained from voltage-clamp electrophysiology of HEK293T cells loaded with 100 nM VF. Lifetime listed here is the average 0 mV lifetime from the electrophysiology calibration. % Δτ/τ is the percent change in lifetime corresponding to a 100 mV step from −60 mV to +40 mV. Lifetime sample sizes: fluorescein 25, erythrosin B 25, VF2.1.Cl 17, VF2.0.Cl 17. For lifetime standards, each measurement was taken on a separate day. VF2.1.Cl data in HEK293T is duplicated in Figure 2—source data 1. Values are tabulated as mean ± SEM.
- https://doi.org/10.7554/eLife.44522.009
-
Figure 1—source data 3
Comparison of optical approaches to absolute Vmem determination in HEK293T cells.
Data are compiled from Figure 1 (VF-FLIM, this work), Figure 1—figure supplement 4 (CAESR; Brinks et al., 2015), and Figure 1—figure supplement 5 (Di-8-ANEPPS; Zhang et al., 1998). All data were obtained by simultaneous whole cell voltage clamp electrophysiology and optical recording in HEK293T (VF-FLIM n = 17 cells, CAESR n = 9, di-8-ANEPPS n = 16). Calculation of intra and inter cell accuracies are performed via root-mean-square deviation (RMSD) and discussed in detail in the Methods (see Resolution of VF-FLIM…). Regions of interest were chosen at the plasma membrane in all cases. Di-8-ANEPPS data are presented as the ratio of signal obtained with blue excitation to signal obtained with green excitation (R, see Materials and methods) and are not normalized to the 0 mV R.
- https://doi.org/10.7554/eLife.44522.010
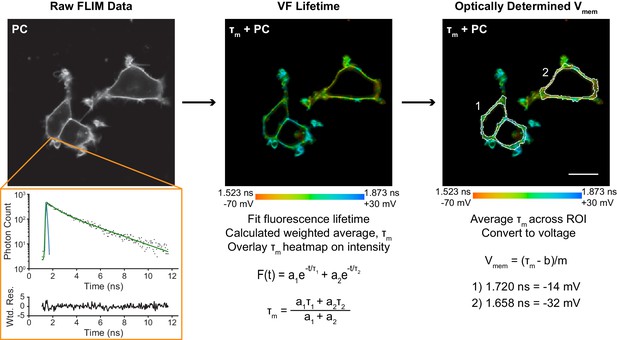
Overview of data processing to obtain membrane potential recordings from fluorescence lifetime.
Time-correlated photon data (black dots, first panel) collected at each pixel were fit to an exponential decay model (green) with iterative reconvolution of the instrument response function (IRF, blue). The two components of the fluorescence lifetimes were converted to a weighted average (middle panel). Cell membranes (white outlines) were identified, and τm was averaged within each of these regions of interest (ROIs, right panel). These lifetimes were then converted to voltage via a previously determined lifetime-Vmem standard curve with slope m and y-intercept b. Additional details of this process are provided in the Methods. Wtd. Res.: weighted residuals of the fit, τm: weighted average fluorescence lifetime, PC: photon count. τm + PC represents an overlay of the lifetime data (color heat map) onto the photon count image. Pixels that appear black in τm + PC images were below the required photon count threshold for fitting lifetime data; PC only images show photon counts without any such thresholds applied. Scale bar is 20 µm.
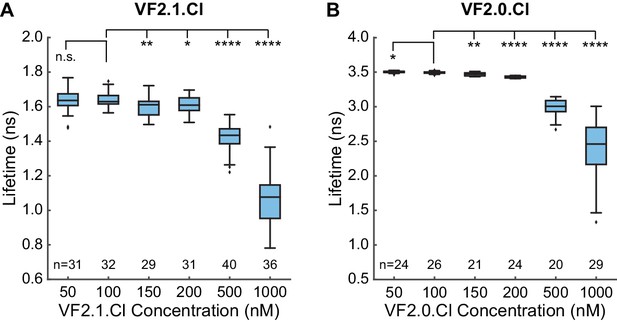
Concentration dependence of VoltageFluor lifetimes in HEK293T cells.
Changes in lifetime arising from addition of a range of concentrations of (A) VF2.1.Cl or (B) voltage-insensitive control VF2.0.Cl in HEK293T cells. Biexponential fit models were used for all VF2.1.Cl concentrations and 1 μM VF2.0.Cl; a monoexponential model was used for all other VF2.0.Cl concentrations. Box plots represent the interquartile range, with whiskers and outliers determined with the Tukey method. Sample sizes indicate number of cell groups. Data were obtained over 2 to 4 different days from a total of 3 or 4 coverslips at each concentration. Asterisks indicate significant differences between the indicated concentration and the VF concentration used for electrophysiology experiments (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test).
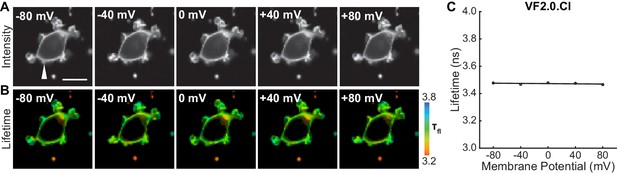
VF2.0.Cl lifetime does not depend on membrane potential.
(A) Photon count and (B) lifetime images of a single HEK293T cell loaded with 100 nM VF2.0.Cl, with the membrane potential held at the indicated value via whole-cell voltage clamp electrophysiology. White arrow indicates patch pipette. Scale bar is 20 μm. (C) Quantification of images shown in (B) for this individual cell. Black line is the line of best fit.
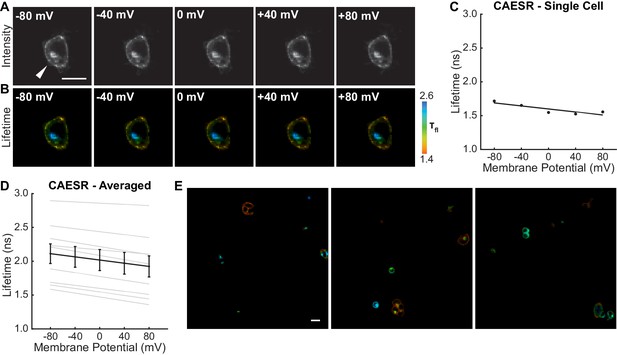
The GEVI CAESR shows variable lifetime-voltage relationships.
(A) Photon count and (B) lifetime images of a HEK293T cell expressing CAESR and held at the indicated Vmem with voltage-clamp electrophysiology. White arrow indicates voltage-clamped cell. (C) Lifetime-Vmem relationship from the cell in (B), based on a single fit from combined fluorescence decays of all pixels in the cell membrane at each potential (see Materials and methods). Points indicate recordings at a given potential; solid line is line of best fit. (D) Evaluation of VF2.1.Cl lifetime-voltage relationships in many individual CAESR-expressing HEK293T cells. Gray lines represent linear fits on individual cells. Black line is the average fit across all cells (n = 9). (E) Representative lifetime images of CAESR in HEK293T cells. Scale bars represent 20 μm.
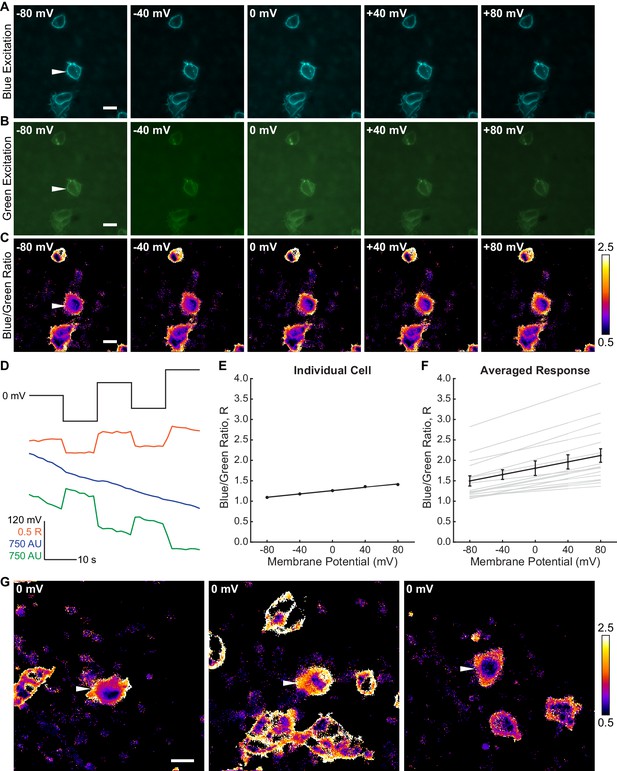
Ratiometric Vmem determinations with Di-8-ANEPPS in HEK293T cells.
Different fluorescence excitation wavelengths (blue and green) were used to generate ratiometric images; emission wavelengths were constant throughout the experiment. Details of epifluorescence ratiometric imaging and data processing are described in the Materials and methods Simultaneous ratiometric imaging and whole cell patch clamp electrophysiology were used to determine the ratio between the blue and green excitation channels (blue/green ratio, R). Representative images of (A) blue-excited signal, (B) green-excited signal and (C) R of a HEK293T cell held at the indicated membrane potential with whole cell voltage clamp electrophysiology. White arrow indicates voltage clamped cell. Differences in background brightness between images are attributable to photobleaching of the probe; images were acquired in the order: 0,–80, +40,–40, +80 mV. (D) Time course of the voltage step protocol used for each cell (black line, top), along with a representative response in the fluorescence ratio (orange), blue excited fluorescence intensity (blue) and green excited fluorescence intensity (green). AU, arbitrary units. (E) R at various potentials for an individual HEK293T cell (black dots), along with an R-Vmem line of best fit (black line). Values shown are the average of all ratio images from a particular cell at a given potential, excluding the first and last ratio image taken for each Vmem value. Data shown in (D) and (E) are from the cell depicted in (A)-(C). (F) Aggregated lines of best fit (gray) for R-Vmem calibrations on multiple HEK293T cells. Average response is shown in black; ratios at each potential are depicted as mean ± SEM (n = 16 cells). (G) Additional representative R images of Di-8-ANEPPS in HEK293T cells. The cell indicated with the white arrow is held at 0 mV with electrophysiology; other cells are unperturbed. Images represent one frame at the given voltage and are not averaged. Regions of interest (ROIs) were drawn around the membrane only, avoiding both the internalized dye signal and the artificially high ratios just outside the cells (which appear as white edges) as much as possible. Scale bars represent 20 µm.
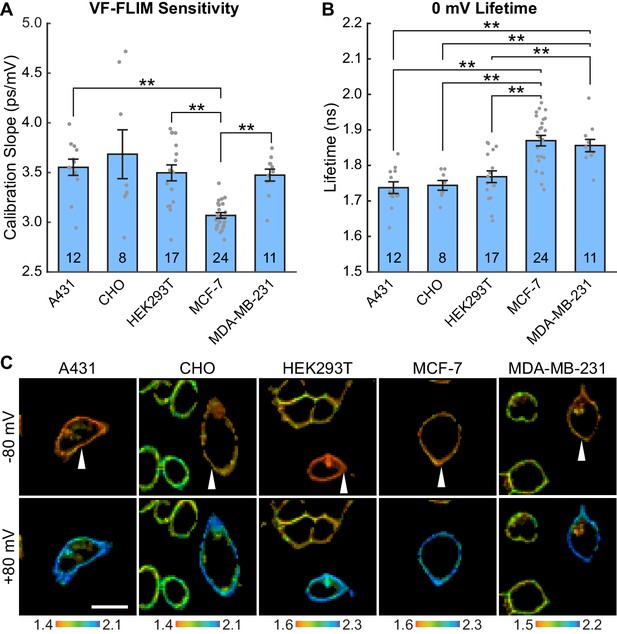
VF-FLIM is a general and portable method for optically determining membrane potential.
VF2.1.Cl lifetime-voltage relationships were determined with whole cell voltage clamp electrophysiology in five cell lines. (A) Slopes of the linear fits for single cell lifetime-voltage relationships, shown as mean ± S.E.M. Gray dots indicate results from individual cells. Statistically significant differences exist between groups (One-way ANOVA with Welch’s correction: F(4, 23.07)=18.12, p<0.0001). Data were tested for normality (Shapiro-Wilk test, p>0.05 for all cell lines) and homoscedasticity (Levene’s test on the median, F(4,67) = 5.07, p=0.0013). ** indicates p<0.01; if significance is not indicated, p>0.05 (Games-Howell post hoc test). (B) 0 mV reference point of linear fits for the lifetime-voltage relationship, shown as mean ± S.E.M. Gray dots indicate results from individual cells. Significant differences exist between groups (One-way ANOVA: F(4, 67)=14.43, p<0.0001). Data were tested for normality (Shapiro-Wilk test, p>0.05 for all cell lines) and homoscedasticity (Levene’s test on the median, F(4,67) = 1.29, p=0.28). ** indicates p<0.01; if significance is not indicated, p>0.05 (Tukey-Kramer post hoc test). (C) Representative lifetime-intensity overlay images for each cell line with the indicated cells (white arrow) held at −80 mV (top) or +80 mV (bottom). Lifetime scales are in ns. Scale bar is 20 μm.
-
Figure 2—source data 1
Lifetime-Vmem standard curves for VF2.1.Cl lifetime in various cell lines.
Whole-cell voltage-clamp electrophysiology was used to determine the relationship between VF2.1.Cl lifetime and membrane potential in five different cell lines. Parameters of this linear model are listed above. The %Δτ/τ is the percent change in the lifetime observed for a voltage step from −60 mV to +40 mV. The intra-cell RMSD represents the accuracy for quantifying voltage changes in a particular cell (see Materials and methods). The inter-cell RMSD represents the expected variability in single-trial absolute Vmem determinations. Sample sizes: A431 12, CHO 8, HEK293T 17, MCF-7 24, MDA-MB-231 11. All values are tabulated as mean ± SEM.
- https://doi.org/10.7554/eLife.44522.018
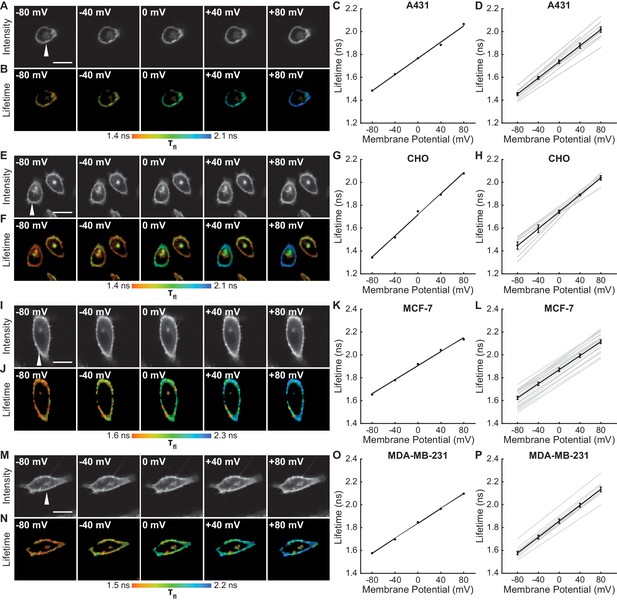
VoltageFluor lifetime reports voltage in diverse cell lines.
(A) Representative photon count and (B) lifetime images of a VF2.1.Cl in A431 cells with Vmem held at the indicated value with voltage-clamp electrophysiology. A431 cells were not serum starved for these experiments. (C) Quantification of the images in (B), with the line of best fit for this single trial. (D) Lines of best fit for the lifetime-Vmem relationships of 12 A431 cells (gray lines). Average lifetime at each potential is shown as mean ± SEM, with the average line of best fit in black. (E)-(H) Lifetime-Vmem standard curve determination in CHO cells (n = 8). (I)-(L) Lifetime-Vmem standard curve determination in MCF-7 cells (n = 24). (M)-(P) Lifetime-Vmem standard curve determination in MDA-MB-231 cells (n = 11). VF2.1.Cl concentration was 100 nM in all cases. White arrows indicates the voltage-clamped cell. Scale bars are 20 μm..
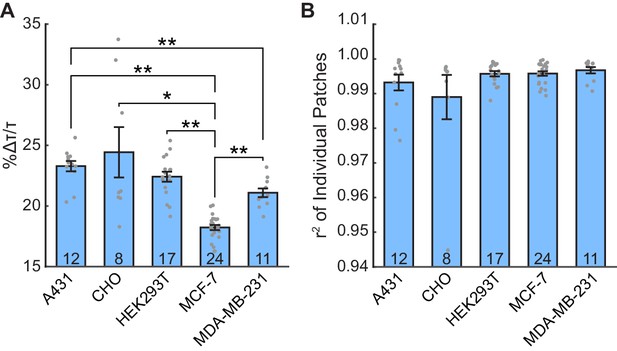
Additional parameters of linear lifetime-voltage standard curves.
(A) Percent change in VF2.1.Cl lifetime per 100 mV change in voltage, relative to the lifetime at −60 mV. Significant differences exist between cell lines (one-way ANOVA with Welch’s correction, F(4,24.08) = 41.75, p<0.0001). Data were tested for normality (Shapiro-Wilk test, p>0.05 for all cell lines) and homoscedasticity (Levene’s test on the median, F(4,67) = 5.74, p=0.00049). Asterisks indicate statistically significant differences (*p<0.05, **p<0.01, Games-Howell post hoc test). (B) Correlation coefficients (r2) for the lines of best fit of VF2.1.Cl lifetime versus membrane potential. No significant differences exist in r2values between cell lines (Kruskal-Wallis test, H = 3.20, 4 degrees of freedom, p=0.53). Data were tested for normality (Shapiro-Wilk test, p<0.05 for 4 of 5 cell lines) and homoscedasticity (Levene’s test on the median, F(4,67) = 1.55, p=0.20). In both (A) and (B), data are shown as mean ± S.E.M., with gray dots indicating values from individual patches.
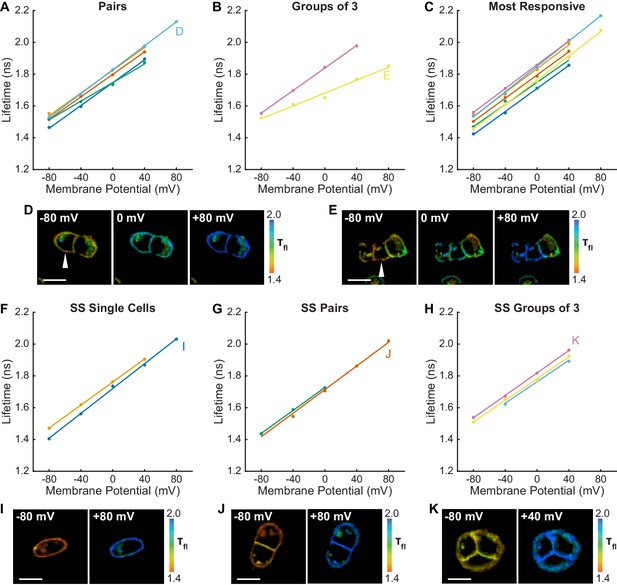
Relationship between lifetime and membrane potential extends to groups of cells and across culture conditions.
Electrophysiological calibration of lifetime was performed on small groups of A431 cells and on serum starved (SS) A431 cells to verify that the Vmem-lifetime standard curves for a given cell line are generalizable across many cellular growth conditions. For all graphs, each line represents a group of cells. Letters on the graphs indicate the subfigure where images from that recording are shown. (A) Lifetime-voltage relationships in cell pairs, in which only one cell was directly controlled with voltage-clamp electrophysiology. (B) Lifetime-voltage relationships in groups of three cells, in which only one cell was directly controlled with voltage-clamp electrophysiology. (C) Lifetime for the most responsive cell from pairs and groups of three in (A) and (B). Line color codes are maintained from (A) and (B). (D, E) Representative lifetime images from (A) and (B) respectively. White arrow indicates cell directly controlled with electrophysiology. (F) Lifetime-voltage relationship in SS single cells, (G) pairs, and (H) groups of three cells. (I)-(K) Representative images from (F)-(H). Scale bars are 20 μm.
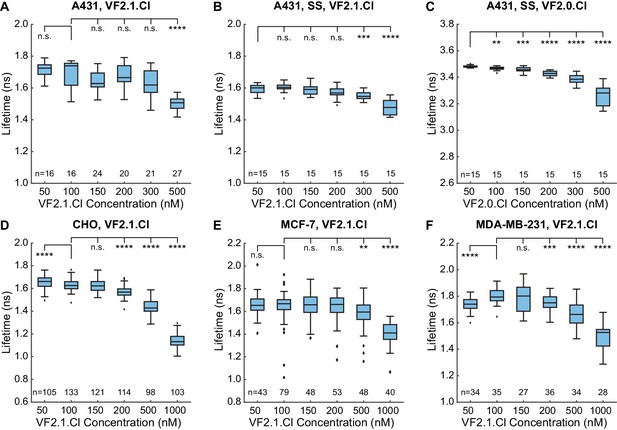
Concentration dependence of VoltageFluor lifetime in four cell lines.
A431 cells were analyzed with VF2.1.Cl both in (A) full serum and (B) serum-starved conditions. (C) VF2.0.Cl in serum-starved A431 cells. (D) VF2.1.Cl in CHO cells. (E) VF2.1.Cl in MCF-7 cells. (F) VF2.1.Cl in MDA-MB-231 cells. All VF2.1.Cl data were fit with a biexponential model, and all VF2.0.Cl data were fit with a monoexponential model. Box plots represent the interquartile range, with whiskers and outliers determined with the Tukey method. Sample sizes indicate number of cell groups. Data were acquired over 2 to 4 different days from a total of 3 or 4 coverslips at each concentration. Asterisks indicate significant differences between the indicated concentration and the VF concentration selected for additional experiments (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test).
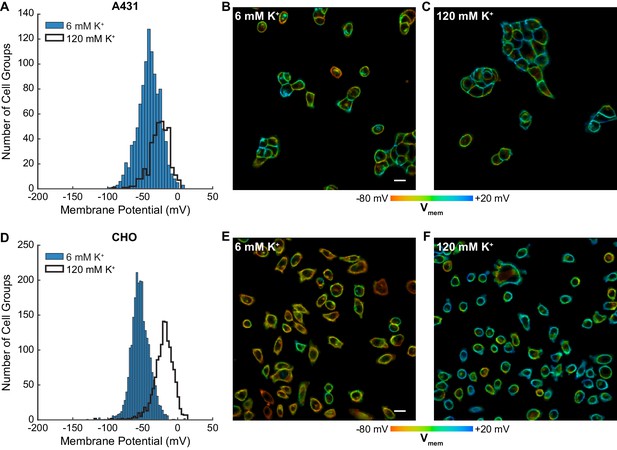
Rapid optical profiling of Vmem at rest and in high extracellular K+.
Fluorescence lifetime images of cells incubated with 100 nM VF2.1.Cl were used to determine Vmem from previously performed electrophysiological calibration (Figure 2). (A) Histograms of Vmem values recorded in A431 cells incubated with 6 mM extracellular K+ (commercial HBSS, n = 1056) or 120 mM K+ (high K+ HBSS, n = 368). (B) Representative lifetime image of A431 cells in 6 mM extracellular K+. (C) Representative lifetime image of A431 cells in 120 mM extracellular K+. (D) Histograms of Vmem values observed in CHO cells under normal (n = 2410) and high K+ (n = 1310) conditions. Representative lifetime image of CHO cells in (E) 6 mM and (F) 120 mM extracellular K+. Histogram bin sizes were determined by the Freedman-Diaconis rule. Intensities in the lifetime-intensity overlay images are not scaled to each other. Scale bars, 20 μm.
-
Figure 3—source data 1
Vmem measurements made with VF-FLIM agree with previously reported values.
Comparison of optically-determined resting membrane potential values (in millivolts) and previously reported values. This table summarizes data presented in Figure 3 and Figure 3—figure supplement 1. Optically determined membrane potentials were calculated from lifetime-Vmem standard curves (Figure 2—source data 1). For tabulated literature values, measures of error and central tendency were used from the original publication. In some cases, none were given or only ranges were discussed. The mean of the reported ephys values is the mean of the values listed here. Sample sizes for resting and elevated K+, respectively: A431 1056, 368; CHO 2410, 1310; HEK293T 1613, 520; MCF-7 1259, 681; MDA-MB-231 1840, 558.
- https://doi.org/10.7554/eLife.44522.022
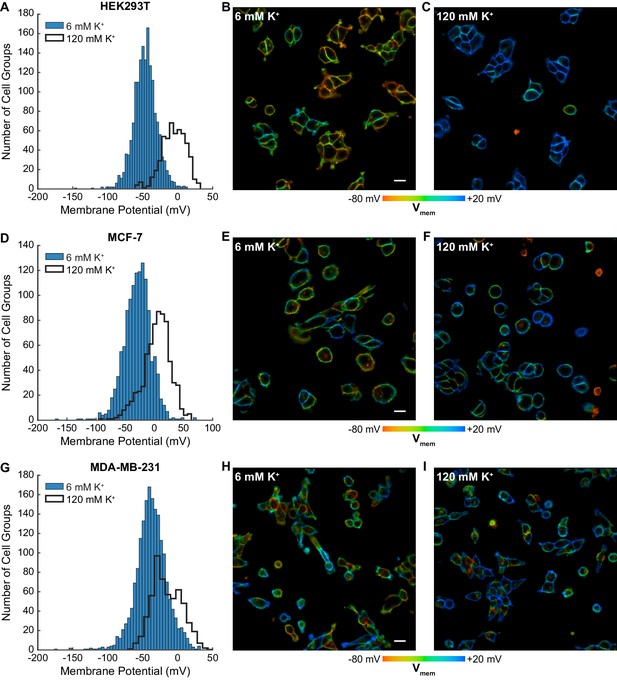
Optically recorded Vmem distributions in HEK293T, MCF-7 and MDA-MB-231 cells.
Fluorescence lifetime images of cells incubated with 100 nM VF2.1.Cl were used to determine Vmem from previously performed electrophysiological calibration (Figure 2). (A) Histograms of Vmem values recorded in HEK293T cells incubated with 6 mM extracellular K+ (commercial HBSS, n = 1613) or 120 mM K+ (high K+ HBSS, n = 520). (B) Representative lifetime image of HEK293T cells with 6 mM extracellular K+. (C) Representative lifetime image of HEK293T cells in 120 mM extracellular K+. (D) Histograms of Vmem values observed in MCF-7 cells under normal (n = 1259) and high K+ (n = 681) conditions. Representative lifetime images of MCF-7 cells in (E) 6 mM and (F) 120 mM extracellular K+. (G) Histograms of Vmem values observed in MDA-MB-231 cells under normal (n = 1840) and high K+ (n = 558) conditions. Representative lifetime images of MDA-MB-231 cells in (H) 6 mM and (I) 120 mM extracellular K+. Histogram bin sizes were determined by the Freedman-Diaconis rule. Intensities in the lifetime-intensity overlay images are not scaled to each other. Scale bars, 20 μm.
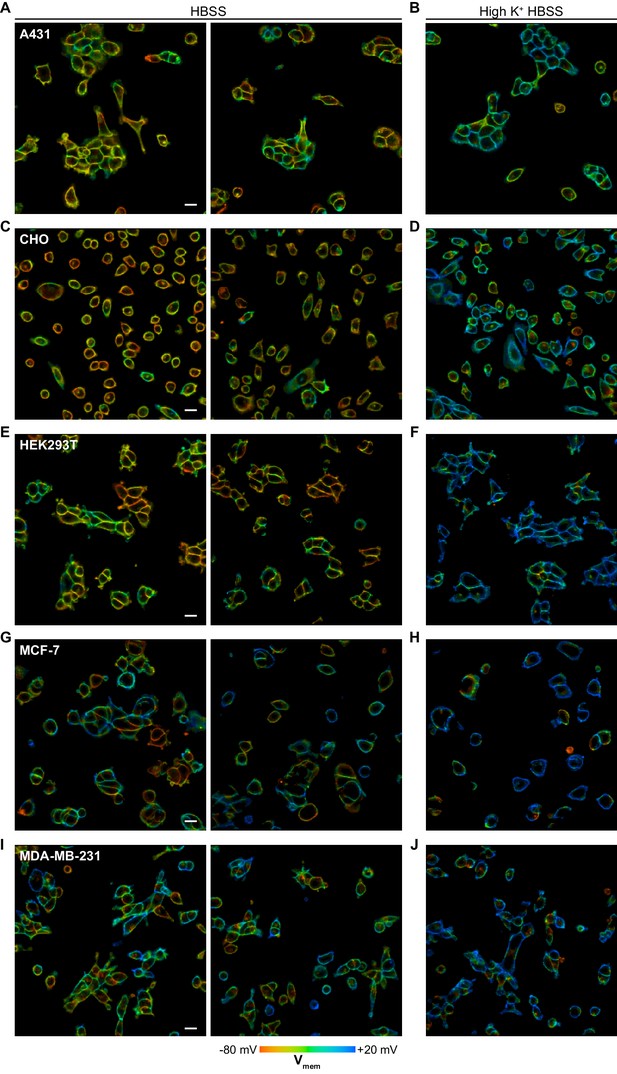
Representative images of cultured cell resting membrane potential.
Representative VF-FLIM images of cells in standard imaging buffer (HBSS, 6 mM extracellular K+) and high K+ imaging buffer (high K+ HBSS, 120 mM extracellular K+). Membrane potential was calculated per cell group; analyses of pixel by pixel differences in lifetime fall beyond the resolution limit of the VF-FLIM calibrations in this work. Images depict A431 cells in (A) HBSS and (B) high K+ HBSS, CHO cells in (C) HBSS and (D) high K+ HBSS, HEK293T cells in (E) HBSS and (F) high K+ HBSS, MCF-7 cells in (G) HBSS and (H) high K+ HBSS, and MDA-MB-231 cells in (I) HBSS and (J) high K+ HBSS.
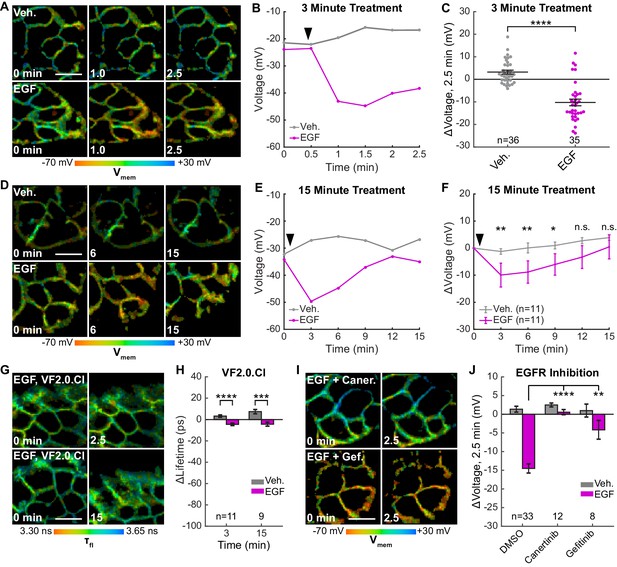
EGFR-mediated receptor tyrosine kinase activity produces a transient hyperpolarization in A431 cells.
(A) Representative VF-FLIM time series of A431 cells treated with imaging buffer vehicle (top) or 500 ng/mL EGF (80 nM, bottom). (B) Quantification of images in (A), with Vehicle (Veh.)/EGF added at black arrow. (C) Aggregated responses for various trials of cells treated with vehicle or EGF. (D) Lifetime images of longer-term effects of vehicle (top) or EGF (bottom) treatment. (E) Quantification of images in (D). (F) Average response of cells over the longer time course. (G) Images of VF2.0.Cl (voltage insensitive) lifetime before and after EGF treatment. No τfl change is observed 2.5 (top) or 15 min (bottom) following EGF treatment. (H) Average VF2.0.Cl lifetime changes following EGF treatment. VF2.0.Cl graphs and images are scaled across the same lifetime range (350 ps) as VF2.1.Cl plots and images. The small drift observed would correspond to 2–4 mV of voltage change in VF2.1.Cl lifetime. (I) Lifetime images of A431 cells before and after EGF addition, with 500 nM canertinib (top) or 10 μM gefitinib (bottom). (J) Voltage changes 2.5 min after EGF addition in cells treated with DMSO (vehicle control) or an EGFR inhibitor. Scale bars are 20 μm. (C,F,H): Asterisks indicate significant differences between vehicle and EGF at that time point. (J): Asterisks reflect significant differences between EGF-induced voltage responses with DMSO vehicle or an EGFR inhibitor (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-tailed, unpaired, unequal variances t-test).
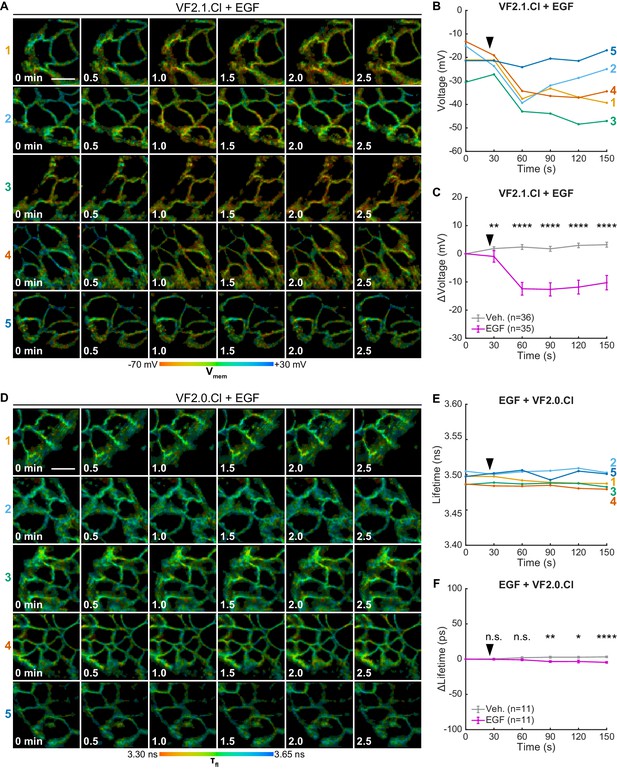
Individual VF-FLIM recordings of A431 EGF response.
(A) Representative 3 min VF-FLIM recordings of A431 cells loaded with 50 nM VF2.1.Cl. 500 ng/mL EGF was added 30 s into the time series (black arrow). (B) Quantification of the images in (A), with a single trace per image series shown. (C) Average voltage change in A431 cells following the addition of imaging buffer vehicle (gray) or EGF (purple). (D) Control VF2.0.Cl (not voltage sensitive, 50 nM) images of A431 cells treated as in (A). Images are scaled across the same amount of lifetime space (350 ps) as the VF2.1.Cl images. (E) Quantification of the images in (D). (F) Average VF2.0.Cl lifetime change seen in A431 cells following the addition of imaging buffer vehicle (gray) or EGF (purple) in A431 cells. Graph is scaled across the same amount of lifetime space as the VF2.1.Cl data in (C). Asterisks indicate significant differences between vehicle and EGF treated cells at a given time point (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test). Scale bars are 20 μm.
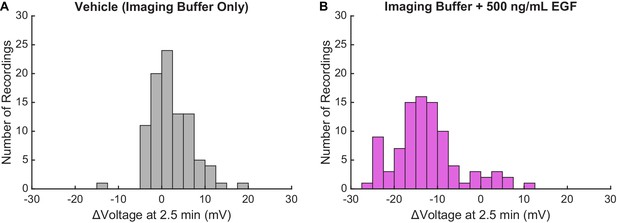
Membrane potential changes in A431 cells 2.5 min after EGF treatment.
Comparison of Vmem changes observed in A431 cells 2.5 min after treatment with (A) imaging buffer vehicle or (B) 500 ng/mL EGF. Data shown here are compiled from Figure 4C and Figure 5A to provide a sense of overall distribution of the responses. Each recording contained a single group of approximately 5 to 10 cells. Sample sizes (number of recordings): Vehicle 93, EGF 92.
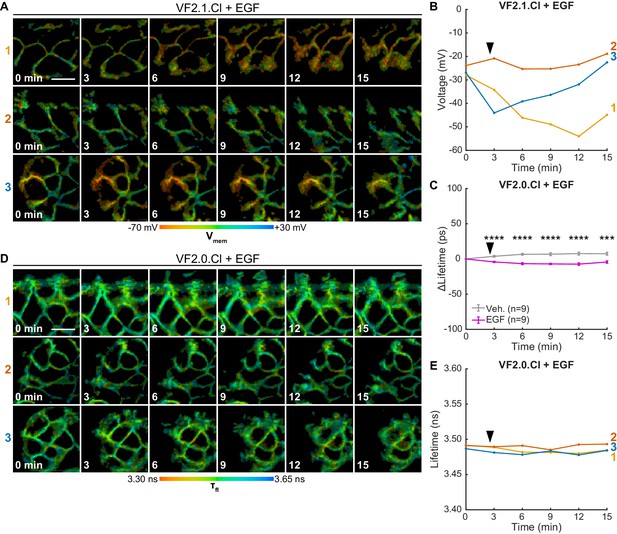
VF-FLIM reports A431 Vmem changes over 15 min.
(A) Representative longer term VF-FLIM recordings of A431 cells loaded with 50 nM VF2.1.Cl. 500 ng/mL EGF was added 30 s into the time series. (B) Quantification of the images in (A), with a single trace per image series shown. (C) Control VF2.0.Cl (not voltage sensitive, 50 nM) images of A431 cells treated as in (A). Images are scaled across the same total lifetime range (350 ps) as the VF2.1.Cl images. (D) Quantification of the recordings in (C). (E) Average VF2.0.Cl lifetime change seen in A431 cells following the addition of imaging buffer vehicle (gray) or EGF (purple). Asterisks indicate significant differences between vehicle and EGF treated cells at a given time point (***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test). Scale bars are 20 μm.
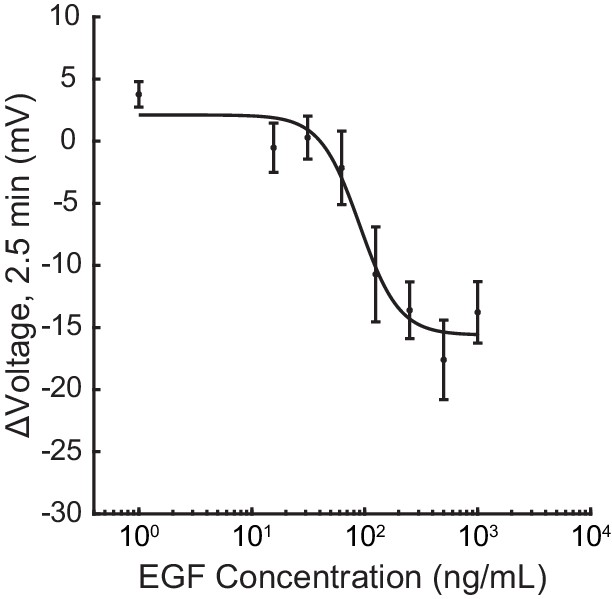
Dose-response relationship of A431 voltage response to EGF.
Data were fit to a four-parameter logistic function to obtain an EC50 of 90 ng/mL (95% CI: 47–130 ng/mL). Response to each EGF concentration is shown as mean ± SEM of 6 or 7 recordings (one group of 5–10 cells per recording).
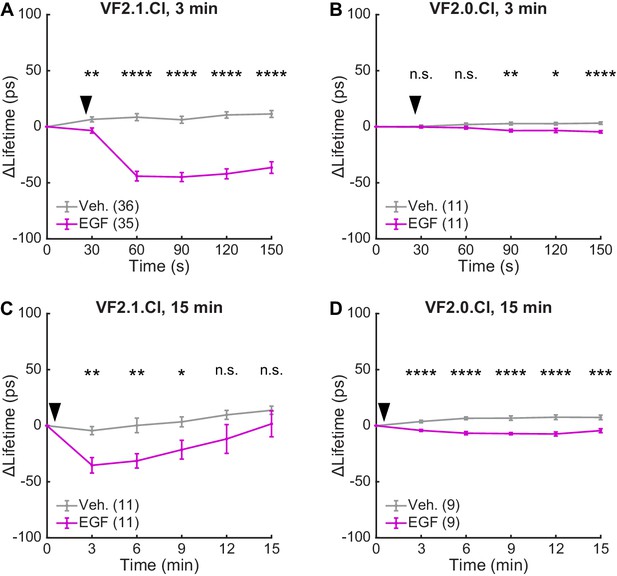
Effect sizes of VF2.1.Cl and VF2.0.Cl response to EGF treatment.
Average lifetime changes observed in A431 cells following the addition (black arrow) of imaging buffer vehicle (gray) or 500 ng/mL EGF (purple). (A) Cells incubated with 50 nM VF2.1.Cl and imaged for 3 min. (B) Cells incubated with 50 nM VF2.0.Cl (not voltage sensitive) and imaged for 3 min. (C) Cells incubated with 50 nM VF2.1.Cl and imaged intermittently for 15 min. (D) Cells incubated with 50 nM VF2.0.Cl (not voltage sensitive) and imaged intermittently for 15 min. Data are reproduced from Figure 4, Figure 4—figure supplement 1, and Figure 4—figure supplement 3, but here data are scaled in units of lifetime rather than voltage for facile comparison. Data are shown as mean ± SEM for the indicated number of recordings (one group of 5–10 cells per recording). Asterisks indicate significant differences between vehicle and EGF treated cells at a given time point (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test).
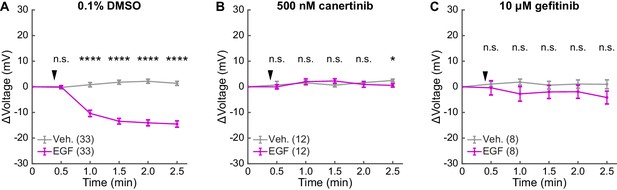
EGFR inhibitors abolish voltage response to EGF in A431 cells.
Average Vmem changes following the addition (black arrow) of imaging buffer vehicle (gray) or 500 ng/mL EGF (purple) to A431 cells pre-treated with the indicated drug or DMSO vehicle. 2.5 min time points from this data are shown elsewhere (Figure 4J); entire time series are shown here. Data are presented as mean ± SEM for the indicated number of recordings (one group of 5–10 cells per recording). Asterisks indicate significant differences between vehicle and EGF treated cells at a given time point (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test).
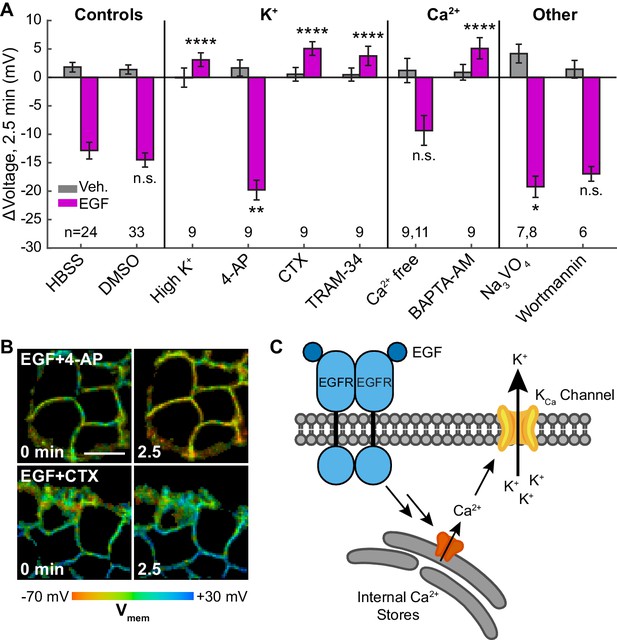
EGF-induced hyperpolarization is mediated by a Ca2+ activated K+ channel.
(A) Comparison of the Vmem change 2.5 min after EGF addition in cells incubated in unmodified imaging buffer (HBSS) or in modified solutions. (B) Lifetime images of A431 cells treated with 4-AP or CTX. (C) Model for membrane hyperpolarization following EGFR activation. Scale bar is 20 μm. Bars are mean ± SEM. Sample sizes listed are (Veh, EGF); where only one number is given, sample size was the same for both. Asterisks reflect significant differences in EGF-stimulated Vmem change between the unmodified control (HBSS or DMSO) and modified solutions (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-tailed, unpaired, unequal variances t-test). DMSO: 0.1% DMSO, high K+: 120 mM K+, 4-AP: 5 mM 4-aminopyridine, CTX: 100 nM charybdotoxin, TRAM-34: 200 nM TRAM-34, Ca2+ free: 0 mM Ca2+ and Mg2+, BAPTA-AM: 10 μM bisaminophenoxyethanetetraacetic acid acetoxymethyl ester, Na3VO4: 100 μM sodium orthovanadate, wortmannin: 1 μM wortmannin.
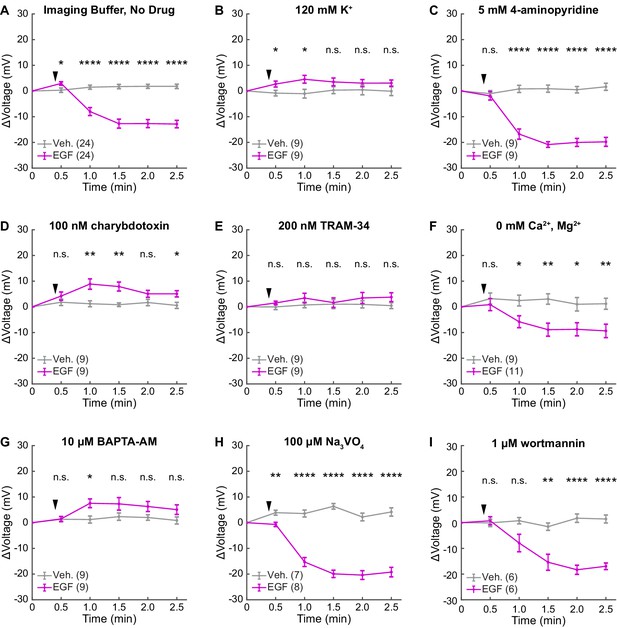
A431 voltage response to EGF with pharmacological intervention.
Average Vmem changes following the addition (black arrow) of imaging buffer vehicle (gray) or 500 ng/mL EGF (purple) to A431 cells pre-treated with the indicated drug or ionic composition change. 2.5 min time points from this data are shown elsewhere (Figure 5); entire time series are shown here to illustrate the time courses of the large hyperpolarizing current and small depolarizing current. Data are shown as mean ± SEM for the indicated number of recordings (one group of 5–10 cells per recording). Asterisks indicate significant differences between vehicle and EGF treated cells at a given time point (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test).
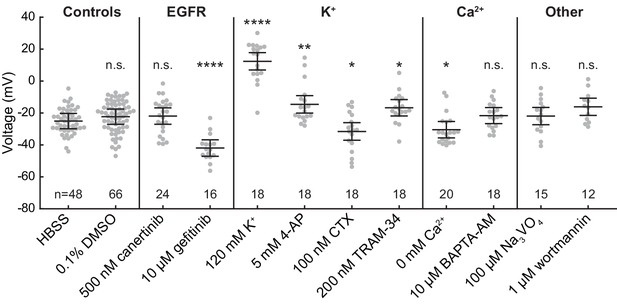
Effects of pharmacological and ionic perturbations on A431 resting membrane potential.
Data are the initial Vmem reference images for recordings used in EGF addition time series. Data are shown as mean ± SEM for the indicated number of images (one group of 5–10 cells per image), and gray dots represent individual images. Asterisks indicate significant differences between the appropriate vehicle (HBSS or 0.1% DMSO) and pharmacology treated cells (n.s. p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-sided, unpaired, unequal variances t-test). CTX = charybdotoxin, 4-AP = 4-aminopyridine, BAPTA-AM = bisaminophenoxyethanetetraacetic acid acetoxymethyl ester.
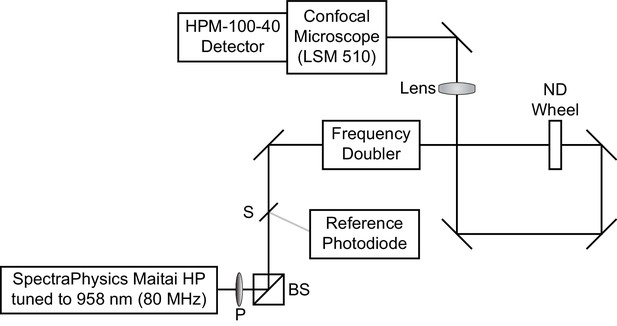
Optical diagram for time correlated single photon counting microscope.
Excitation light was supplied by a Ti:Sapphire laser tuned to 958 nm. A small amount of light was redirected by a beam sampler (S) to a reference photodiode. The remaining light was passed through a frequency doubler to obtain 479 nm excitation light, which entered the LSM510 confocal microscope. A polarizer (P) followed by a polarizing beamsplitter (BS), as well as a neutral density (ND) wheel, allowed control of the amount of light passed to the sample.
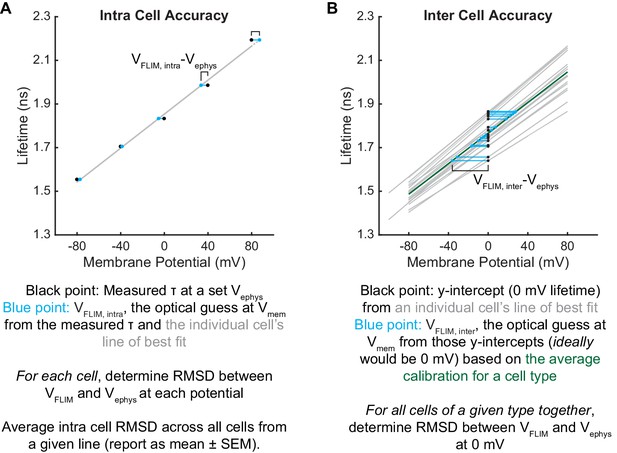
Intra and inter-cell Vmem resolution calculations.
Data are taken directly from Figure 1H,I as an example. (A) Intra cell values are the RMSD between the voltage equivalent of the measured lifetime (VFLIM) and voltage set by electrophysiology (Vephys). VFLIM values are calculated using that particular cell’s line of best fit, so one value is obtained per cell. Here, we present intra cell error as the mean ± SEM of all cells from a given cell line. (B) Inter cell errors are the RMSD between the voltage-equivalent of the 0 mV lifetime for all cells tested from a cell line (VFLIM, determined with the average slope and y-intercept for that cell line) and the ground truth value of 0 mV. Inter-cell accuracy is calculated from all of the calibration data for a cell line, so there is one value per cell line. Black points are experimental y-intercepts and blue points are the VFLIM optical voltage determinations from those lifetimes. Gray lines are lines of best fit for individual cells. Green line in (B) represents the average τfl-Vmem relationship for a cell line.
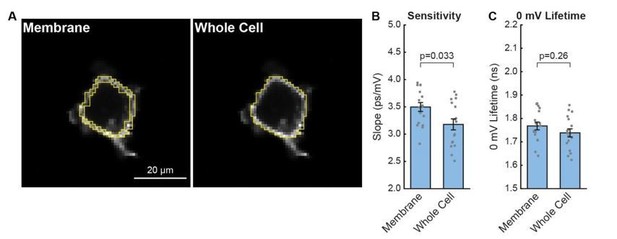
Effect of internal signal on HEK293T VF-FLIM calibrations.
(A) Illustration of different regions of interest (ROIs) used for processing the data. All VF-FLIM analysis in other parts of the manuscript was performed with “membrane” ROIs. Whole cell ROIs include all interior pixels that are above the threshold for lifetime fitting (300 peak photons, see Materials and Methods section). (B) Effect of internal signal on the sensitivity (slope) of the HEK293T τfl-Vmem data shown in Figure 1 of the main text. Gray points are results from individual cells; aggregated data are shown as mean ± SEM of n=17 cells. (C) Effect of internal signal on the 0 mV lifetime (y-intercept) of the lifetime-Vmem calibration. P values for (B) and (C) were determined using paired Student’s t tests.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens, female) | A431 | UC Berkeley Cell Culture Facility | RRID:CVCL_0037 | Cell line maintained in E. Miller lab |
| Cell line (Homo sapiens, female) | HEK293T | UC Berkeley Cell Culture Facility | RRID:CVCL_0063 | Cell line maintained in E. Miller lab |
| Cell line (Homo sapiens, female) | MCF-7 | UC Berkeley Cell Culture Facility | RRID:CVCL_0031 | Cell line maintained in E. Miller lab |
| Cell line (Homo sapiens, female) | MDA-MB-231 | UC Berkeley Cell Culture Facility | RRID:CVCL_0062 | Cell line maintained in E. Miller lab |
| Cell line (Cricetulus griseus, female) | CHO | UC Berkeley Cell Culture Facility | RRID:CVCL_0214 | Cell line maintained in E. Miller lab |
| Recombinant DNA reagent | CAESR, FCK-QuasAR2-Citrine | Addgene, PMID: 25118186 | Addgene:59172, RRID:Addgene_59172 | Developed by Adam Cohen, Harvard University |
| Peptide, recombinant protein | Recombinant human epidermal growth factor (EGF) | PeproTech | Cat#:AF10015500UG | |
| Commercial assay or kit | Lipofectamine 3000 | Thermo Fisher Scientific | Cat#:L3000008 | |
| Commercial assay or kit | QIAprep spin miniprep kit | VWR International | Cat#:27106 | |
| Chemical compound, drug | Sodium orthovanadate | Sigma-Aldrich | CAS:13721-39-6, Cat#:S6508 | Activated before use (Gordon, 1991) |
| Chemical compound, drug | Canertinib | other | CAS:267243-28-7 | Gift from John Kuriyan, UC Berkeley |
| Chemical compound, drug | Gefitinib | Fisher Scientific | CAS:184475-35-2, Cat#:50-101-6270 | |
| Chemical compound, drug | 4-aminopyridine, 4-AP | Sigma-Aldrich | CAS:504-24-5, Cat#:A78403 | |
| Chemical compound, drug | Charybdotoxin, CTX | Sigma-Aldrich | CAS:95751-30-7, Cat#:C7802 | |
| Chemical compound, drug | TRAM-34 | Sigma-Aldrich | CAS:289905-88-0, Cat#:T6700 | |
| Chemical compound, drug | BAPTA-AM, bisamino-phenoxy-ethanetetra-acetic acid acetoxymethyl ester | Fisher Scientific | CAS:126150-97-8, Cat#:50-101-0334 | |
| Chemical compound, drug | wortmannin | Fisher Scientific | CAS:19545-26-7, Cat#:ICN19569001 | |
| Software, algorithm | SPCM | Becker and Hickl | ||
| Other | Di-8-ANEPPS | Thermo Fisher Scientific | CAS:157134-53-7, Cat#:D3167 | |
| Other | VF2.1.Cl | Synthesized in-house (Woodford et al., 2015) | ||
| Other | VF2.0.Cl | Synthesized in-house (Woodford et al., 2015) |
Additional files
-
Source code 1
Global analysis of CAESR fluorescence lifetimes.
- https://doi.org/10.7554/eLife.44522.034
-
Source code 2
Automated thresholding and cell group identification.
- https://doi.org/10.7554/eLife.44522.035
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44522.036





