Ion counting demonstrates a high electrostatic field generated by the nucleosome
Figures
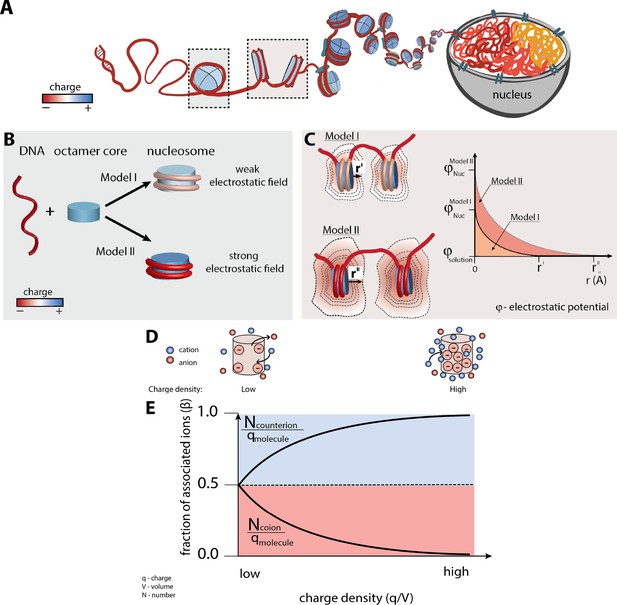
Nucleosome electrostatics: models and implications.
(A) Schematic representation of nucleosome formation and DNA compaction. (B) Models of nucleosome electrostatics. According to Model I, the electrostatic field around the nucleosome is weak due to compensatory electrostatic interactions between the DNA and the positively charged histone octamer. In Model II the electrostatic field remains strong, as has been proposed by theoretical and computational studies. (Schiessel, 2003; Materese et al., 2009; Elbahnsi et al., 2018) (C) Schematic representation of the effect of nucleosome electrostatics on their propensity to compact according to Models I and II. The distance for full screening of electrostatic repulsion is considerably less for Model I (top) than for Model II (bottom; rI vs. rII). (D) Schematic representation of an ion atmosphere around a low charge density (ρ) (left) and a high charge density molecule (right). (E) Fraction of associated counterions (e.g., cations around a negatively charged molecule) and coions (e.g., anions around a negatively charged molecule) within a molecule’s ion atmosphere as a function of the molecule’s charge density (assuming a uniform charge distribution for simplicity). The magnitude of the electrostatic field correlates with the charge density of the molecule: the higher the charge density, the larger magnitude of the electrostatic field and stronger counterion attraction, as depicted by the cartoons in part (D).
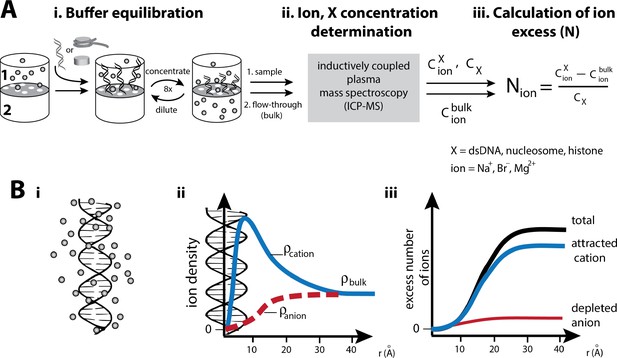
Experimental characterization of the ion atmosphere around macromolecules by ‘ion counting’.
(A) Schematic of an ‘ion counting’ experiment to quantify the composition of the ion atmosphere around dsDNA, nucleosomes, histones or the histone octamer (Bai et al., 2007; Gebala et al., 2015; Greenfeld and Herschlag, 2009). A detailed description of the ion counting method can be found in the Materials and methods section. Adapted from reference (Gebala et al., 2015). (B): i) Schematic representation of the cation excess surrounding a nucleic acid. ii) Schematic representation of ion density as a function of distance from a nucleic acid estimated by Poisson-Boltzmann (PB) theory; PB predictions adapted from (Giambaşu et al., 2014). Counterions and coions are radially distributed within the ion atmosphere around charged molecules. The highest density of counterions localizes near to the surface of the molecule, where the highest electrostatic potential resides; far from the molecule the density equals that of the bulk solution. Coions give the opposite behavior, with the lowest density near the molecule’s surface due to electrostatic repulsion. iii) Computationally, the number of ions associated (the excess number, (N) with the ion atmosphere is calculated by integrating the excess ion density around the nucleic acid (Equation 4 in Material and methods).
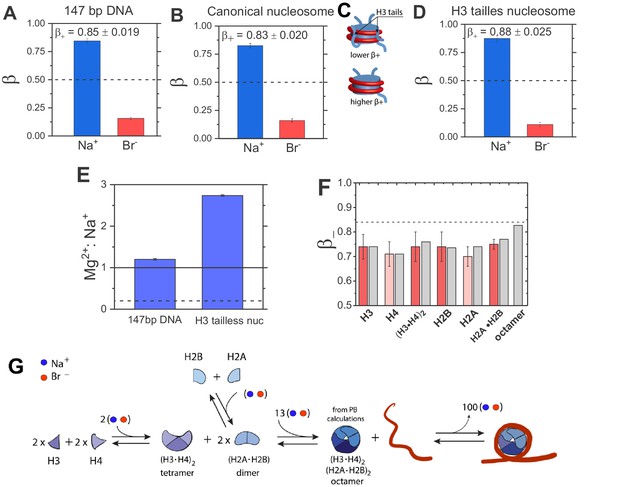
Quantification of electrostatic properties of a free dsDNA, nucleosomes, and histones by ion counting.
(A) Experimentally-determined β coefficients of accumulated Na+ cation (blue bar) and excluded Br– anions (red bar) around 147 bp DNA, (B) canonical nucleosomes, (D) H3 tailless nucleosomes. The β+ values for the canonical and H3-tailless nucleosomes are significantly different based on two-sample t-test, with a p-value of 0.028. Dashed lines represent β = 0.5, the value predicted by the limiting case of Model I. The bulk concentration of NaBr was 10 mM in 2 mM Na-EPPS, pH 7.5 (E) Mg2+ vs. Na+ competition assay for 147 bp DNA and H3 tailless nucleosome. The solid line represents measurements for a model system, a short 24 bp DNA and the dashed line represents Mg2+:Na+ ratio for a low charge density molecule, an equivalent to β = 0.5. (F) β– coefficients for histone proteins and their complexes determined from ion counting results. Experimental results are shown in red bars and theoretical PB calculations are shown in gray bars. Ion counting experiments were carried out at 40 mM NaBr in 2 mM Na-EPPS, pH 7.5, whereas PB calculation were carried out at 40 mM monovalent salt. Dashed line represents the β+ value for 147 bp DNA from Figure 3A. Each data point is the average of two repeats from at least two independent experiments. See Figure 3A—source data 1-7 for raw data. (G) Schematic representation of octamer and nucleosome formation. Based on ion counting results and PB calculations for the histone octamer (Figure 3A—source data 1-7) we estimated the number of ions required at each step of the assembly, shown by the numbers in the assembly scheme. The formation of the octamer complex requires an uptake of ions to attenuate electrostatic repulsion between positively charged histone proteins, whereas ion release accompanies nucleosome formation. An equal number of cations and anions are taken up or released, as required (counterintuitively) to maintain charge neutrality (see Lipfert et al., 2014; Misra and Draper, 2002; Draper, 2004 for explanation).
-
Figure 3—source data 1
Experimentally determined excess number (), the coefficient (the faction of associated cations), and the coefficient (the faction of excluded anions)) for 10 mM NaBr around 147 bp DNA.
- https://doi.org/10.7554/eLife.44993.008
-
Figure 3—source data 2
Experimentally determined excess number (), the coefficient (the faction of associated cations), and the coefficient (the faction of excluded anions) for 10 mM NaBr around canonical nucleosome.
- https://doi.org/10.7554/eLife.44993.009
-
Figure 3—source data 3
Experimentally determined excess number (), the coefficient (the faction of associated cations), and the coefficient (the faction of excluded anions) for 10 mM NaBr around H3 tailless nucleosome.
- https://doi.org/10.7554/eLife.44993.010
-
Figure 3—source data 4
Experimentally determined excess number () for 25 mM NaBr and 2.5 mM MgBr2 around 24 bp DNA, 147 bp DNA and H3 tailless nucleosome.
- https://cdn.elifesciences.org/articles/44993/elife-44993-fig3-data4-v2.pdf
-
Figure 3—source data 5
PB calculation of excess number () and the coefficient (the faction of associated anions), and the coefficient (the faction of excluded cation) for 40 mM NaBr around histone octamer.
- https://doi.org/10.7554/eLife.44993.012
-
Figure 3—source data 6
Experimentally determined excess number () and the coefficient (the faction of associated anions) and the coefficient (the faction of excluded cation) for 40 mM NaBr around histones H3, H4 and (H3·H4)2 tetramer.
- https://doi.org/10.7554/eLife.44993.013
-
Figure 3—source data 7
Experimentally determined excess number () and the coefficient (the faction of associated anions) and the coefficient (the faction of excluded cation) for 40 mM NaBr around histones H2A, H2B and (H2A·H2B) dimer.
- https://doi.org/10.7554/eLife.44993.014
-
Figure 3—source code 1
Ion counting by Poisson Boltzmann.
- https://doi.org/10.7554/eLife.44993.015
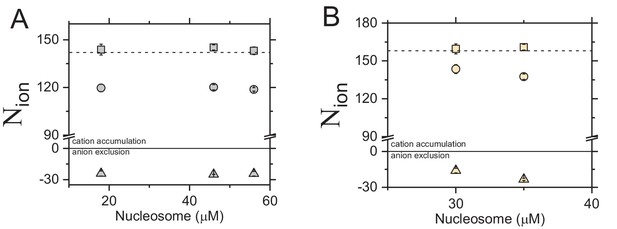
Preferential association of ions (N) around a nucleosome and a function of the molecule concentration.
Accumulation of Na+ (circles) and repulsion of Br— (triangle) around the canonical nucleosome (A) and the H3 tailless nucleosome (B). Sum of cations and anions (total: squares) within the ion atmosphere counterbalances the total charge of the molecule (a dashed line at +144 in A and +160 in B). Each data point is the average of 3–8 independent repeats.
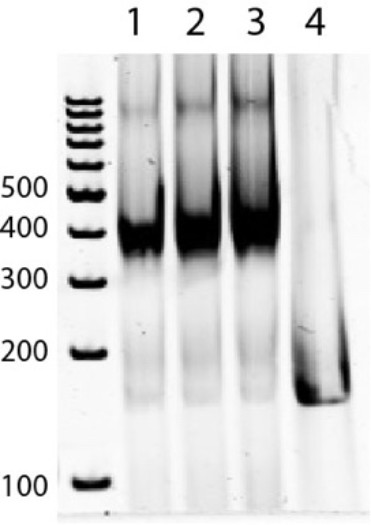
Non-denaturation PAGE (5%) showing nucleosomes before ion counting (1) and after ion counting experiments with 10 mM NaBr (2) and a mixture of 2.5 mM MgBr2 and 25 mM NaBr (3) as well free 147 bp DNA (4).
Most left lane is a dsDNA ladder.
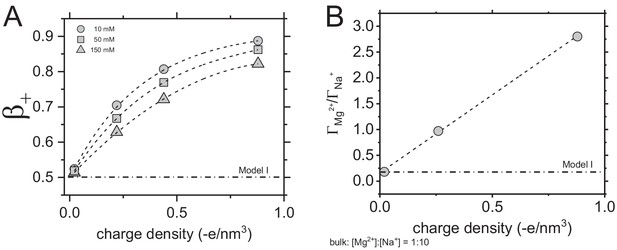
Poisson-Boltzmann (PB) calculations of β+ for Na+ (A) and Mg2+ and Na+ competition (B) as a function of molecule charge density.
PB calculations were carried out for 10, 50, and 150 mM NaCl (A) and Mg2+:Na+ mixture at concentration 1 mM Mg2+ and 10 mM Na+. Attraction of monovalent cations to a negatively charged polyelectrolyte follows exponential dependence on the molecule’s charge density, as shown in (A). Whereas, Mg2+:Na+ ratio increases linearly with the increase of the molecule’s charge density (B). The dash lines are shown to guide the eye and the dash dot lines represent predictions for the Model I (see main text).
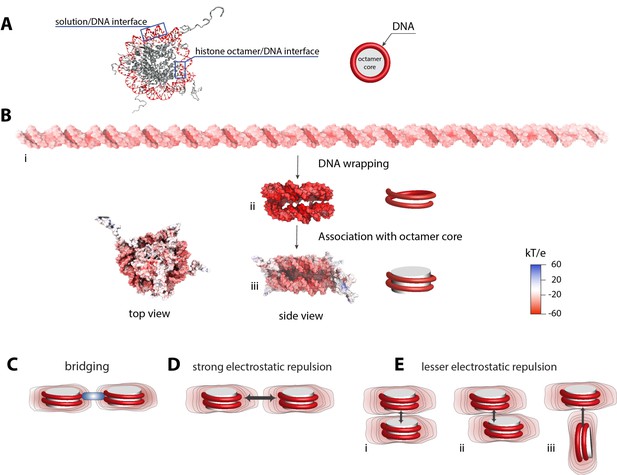
Electrostatic surface potential of a nucleosome and a dsDNA.
(A) Crystal structure of a nucleosome (pdb 1kx5) (Davey et al., 2002). (B) Poisson-Boltzmann calculations of electrostatic surface potential of a dsDNA and a nucleosome (pdb 1kx5); the electrostatic potential ranges from +60 (blue) to −60 (red) kTe−1, which highlights the difference in potential magnitude around the DNA and octamer core. The electrostatic potential mapped to the molecular surfaces was calculated using Adaptive Poisson-Boltzmann Solver (APBS) (Dolinsky et al., 2004) and the figures were rendered with VMD (Humphrey et al., 1996). The electrostatic surface potential of the nucleosomal DNA is largely negative and shown in red. (C)-(E) Schematic representation of electrostatic field lines around a nucleosome and models of internucleosomal interactions. The differences in the electrostatic field as shown in (B) may play an important role in internucleosomal interactions. High electrostatic potential of the nucleosomal DNA disfavors a side-by-side orientation as shown in (D) but introduce the possibility of bridging interactions stabilized by positively charged polyamines and proteins (C, shown in blue). Stacking of nucleosomes with the octamer-to-octamer orientation is energetically more favorable due to weaker electrostatic potentials at the octamer facets as shown in (D).
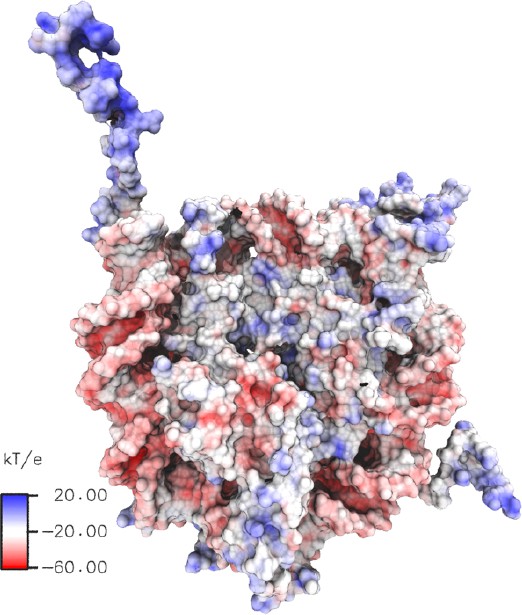
Poisson-Boltzmann calculations of electrostatic surface potential of a nucleosome particle (pdb 1kx5).
The electrostatic potential ranges from +20 (blue) to −60 (red) kTe−1. The electrostatic potential mapped to the molecular surface was calculated using Adaptive Poisson-Boltzmann Solver (APBS) (Dolinsky et al., 2004) and the figure was rendered with VMD (Humphrey et al., 1996).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | BL21(DE3)pLysS | Agilent #200132 | ||
| Recombinant DNA reagent | Histone H2A expression plasmid | Luger et al., 1997, Dyer et al., 2004 | gift from Narlikar Lab | |
| Recombinant DNA reagent | Histone H2B expression plasmid | Luger et al., 1997, Dyer et al., 2004 | gift from Narlikar Lab | |
| Recombinant DNA reagent | Histone H4 expression plasmid | Luger et al., 1997,Dyer et al., 2004 | gift from Narlikar Lab | |
| Recombinant DNA reagent | Histone H3 expression plasmid | Luger et al., 1997, Dyer et al., 2004 | gift from Narlikar Lab | |
| Recombinant DNA reagent | Histone H3-tailless expression plasmid | Luger et al., 1997, Dyer et al., 2004 | gift from Narlikar Lab | |
| Software, algorithm | apbs-pdb2pqr | http://www.poissonboltzmann.org/apbs/ | ||
| Software, algorithm | OriginPro 2017 | https://www.originlab.com |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.44993.018





