Self-capping of nucleoprotein filaments protects the Newcastle disease virus genome
Figures
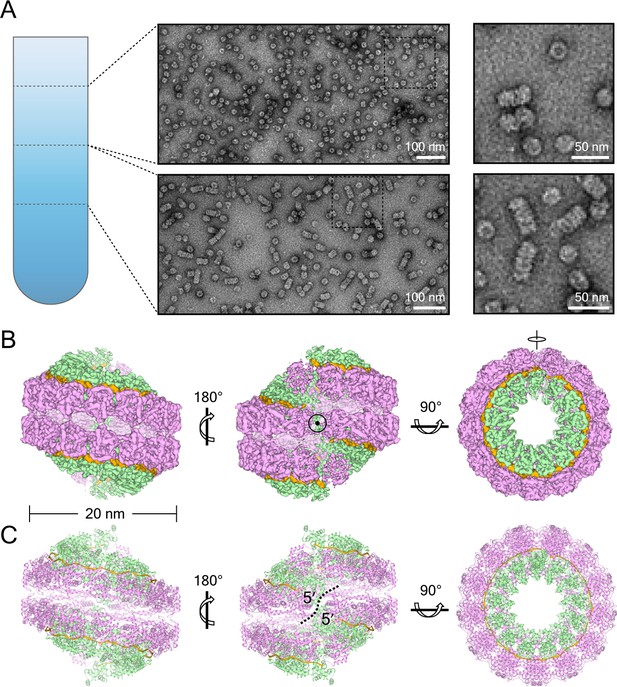
NDV N assembles into clam-shaped structures with two single-strand spirals packing in a back-to-back manner.
(A) The images show negative-stain EM micrographs of the round-shaped structures (top image, upper fraction) and filaments (bottom image, lower fraction) after sucrose-gradient centrifugation (close-ups of the boxed areas are shown on the right). (B) Various views of the 3D reconstruction of the clam-shaped structure of N from the upper fraction. The C2 symmetry axis enforced during reconstruction is indicated in the center view (middle). The NTD, CTD and RNA are colored in pink, green and gold, respectively. (C) Atomic model of the clam-shaped structure of N shown from the same view as in (B) and using the same color code. The two 5′ ends of the enwrapped RNA and the seam between them are labeled in the middle view.
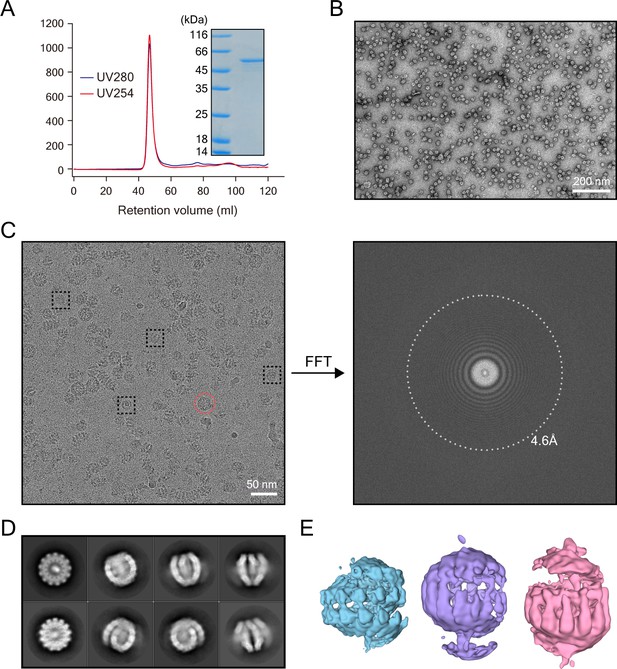
Data analysis of the clam-shaped structure of NDV N.
(A) The size-exclusion chromatography and SDS-PAGE profile of NDV N. The retention volume of N was ~47 ml and the A260/A280 of N was around 1.1. N from the SDS-PAGE gel was of high purity. (B) Negative-stain image of N after gel filtration (enlarged view of negative stain image in Figure 3D). Round-shaped structures are visible with a small portion of filaments of different lengths. (C) A typical cryo-EM micrograph and its Fast Fourier Transform (FFT). The micrograph was taken under a Titan Krios G2 microscope equipped with Gatan K2 summit camera and was motion corrected. Representative particles in different orientations are indicated with dashed black squares. Qβ virus-like particles for improving orientation distribution are shown in the red circle. The direct FFT of the micrograph shows the highest signal frequency at 4.6 Å or higher. (D) Eight typical two-dimensional (2D) classes show a wide range of angular orientations. (E) Three-dimensional (3D) classification of N.

3D reconstruction of the clam-shaped structure of N and resolution estimations.
(A, B) 3D structures after refinement without C2 (A) or with C2 (B) symmetry. The pseudoatomic structures of N and poly-Uracil (poly-U) are docked into the EM map. Two views are shown and the seam is indicated using dashed lines. Dust has not been removed. (C) Angular distribution of the clam-shaped structure with C2 symmetry. The major angular distribution peak and the respective structures are shown. (D) Fourier shell correlation (FSC) curves of clam-shaped structures with or without C2 symmetry, unmasked or masked (masking-effect-corrected FSC) based on 0.143 criteria. The structure with masked C2 symmetry was selected for the following analysis. (E) Local resolution analysis with Resmap shows an overall resolution of ~5 Å. The seam position is indicated with a dashed line in the middle view. The resolution gradually declines from the protomers furthest away from the seam to the protomers closest to the seam (right).

Structural analysis of the clam-shaped structure of NDV N.
(A) The domain organization of NDV N. The NTD and CTD are illustrated in pink and green, respectively. The carboxyl tail is unresolved in the 3D structure and is boxed by a dashed line. (B) Homology modeling of NDV N using a subunit of the PIV5 nucleocapsid (gray) as the template. (C) Docking of one N and poly-U pair into the clam-shaped EM map with RNA shown in gold. (D) Comparison between NDV N in an active state and Nipah N in an inactive state (gray). The transition is of a rotation of the CTD by 24°. (E) Horizontal packing among neighboring N. Each N interacts with its neighboring N with its N-arm and C-arm (boxed in red). (F) RNA enwrapped in the clam-shaped structure and the zoomed-in protomer and RNA. (G) Electrostatic potential distribution of one protomer at pH 7.4. The cleft between the NTD and the CTD is positively charged and binds negatively charged RNA.
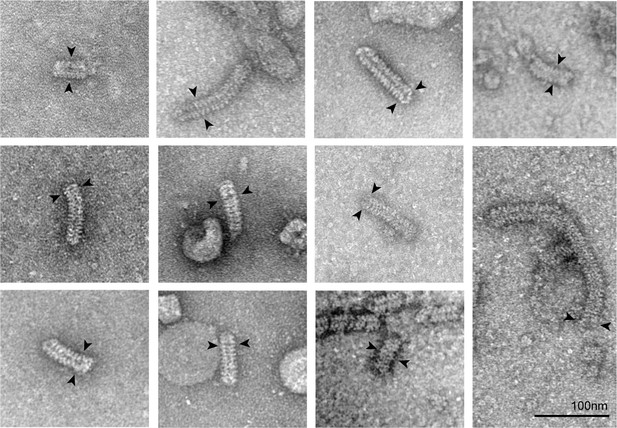
The negative-stain EM of the N–RNA complex isolated from NDV.
The N–RNA complexes are double-headed and the clam-like cores are labeled with arrows.
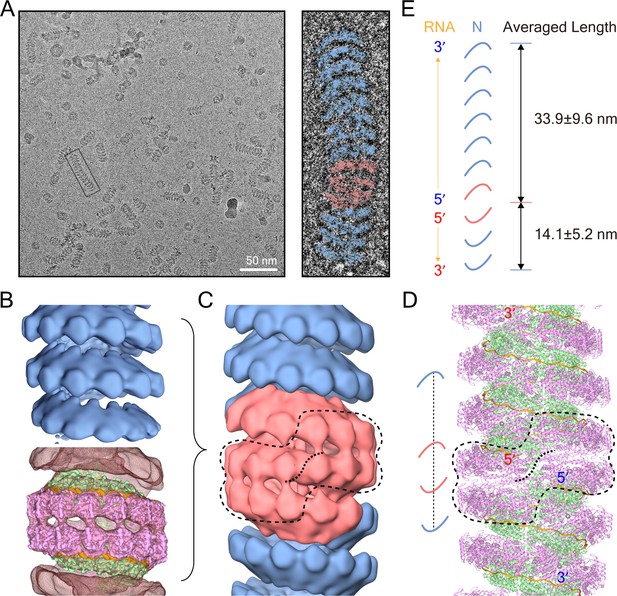
Double-headed filament derived from the clam-shaped structure.
(A) Representative cryo-EM micrograph of the N filament from the lower fraction. One typical double-headed filament was selected, magnified and colored in blue (helical structure) and red (clam-shaped core). (B) 3D reconstructions of the helical structure (top) and the clam-shaped core (bottom). The 4.8 Å clam-shaped structure is docked into the clam-shaped core in the filament. (C) Combination of both helical filaments and the clam-shaped core yields the whole double-headed filament. The position of the clam-shaped core in the composite structure is delineated by the dashed line. (D) Atomic model of the double-headed filament shows the position of the clam-shaped core (dashed line). Corresponding 5′ and 3′ ends from the same RNA are labeled in red and blue. (E) The two helixes in one double-headed filament are of unequal length. The length of each helix is defined as the distance between the helix tip and the center of the clam-shaped core in the cartoon. The length measurements and the RNA direction from 5′ to 3′ are given.
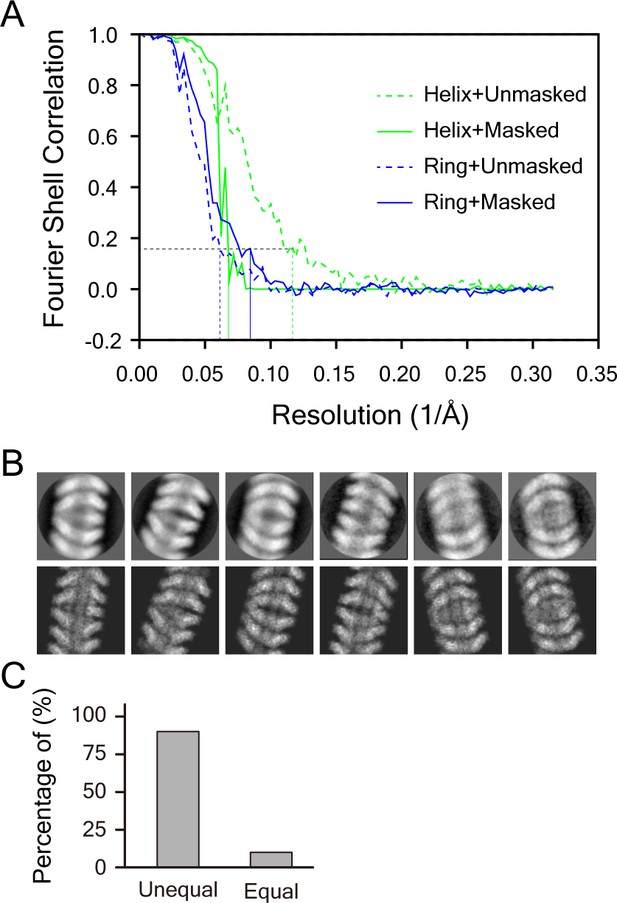
Structural analysis of the double-headed filament.
(A) FSC curves of the helical part and the clam-shaped core based on 0.143 criteria. The final resolutions for the helical part and the clam-shaped core are estimated at 15 Å and 14 Å, respectively. (B) Matching between typical 2D classes (top row) and the respective projections (bottom row, low-pass filtered to 10 Å) of the atomic model of double-headed filaments. (C) Length statistics of two single spirals in one double-headed filament. The length is defined as the distance between the core center and the tip of a single spiral.

Structural analysis of a single-headed filament of the NLoop.
(A) Typical cryo-EM micrograph of the NLoop (enlarged view of Figure 3C). Two representative round-shaped particles were selected (circled) and one 2D averaged class is shown in the top right corner, with 13 protomers recognized. One single-headed filament is marked in the rectangular box and used in further analysis. (B) Power spectrum and 2D classification analysis of a single-headed filament. A single-headed filament is divided into head, middle and tail parts (left), 2D classes of which are shown (middle). Power spectrum analysis of selected filament clearly indicates a 1/60 Å layer line (right). (C) 3D reconstruction of a single-headed filament and the respective pseudo-atomic model. (D) 3D projection of pseudo-atomic model of (C) after a 15 Å low-pass filter with sharp herringbone shape.
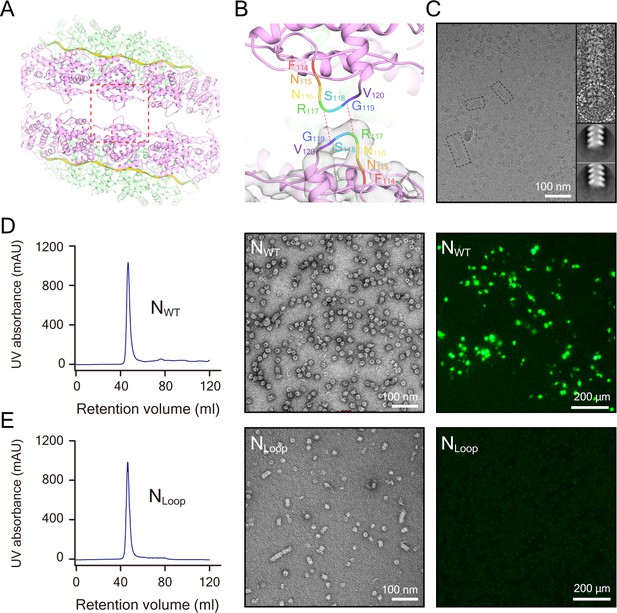
The clam-shaped nucleocapsid is important for the function of the viral genome.
(A) Loop pairs from the vertically adjacent N form the self-capping interface in the clam-shaped structure. Five loop pairs furthest from the seam are shown. Colors are as in Figure 1. (B) View of one loop pair of the clam-shaped structure. Seven residues (114–120) in the upper loop are labeled and the lower loop is docked into the EM density. (C) Raw micrograph of a single-headed helix from the NLoop and the 2D classification of the filament tip (circled). Zoomed-in view of selected raw filaments (examples in dashed boxes) with two typical 2D classes on the tip shown. (D) NWT was able to form double-headed filaments and functioned well in the minigenome assay. The retention volume of NWT in gel filtration chromatography was ~47 ml (left) and the negative-stain image of this fraction consisted of a clam-shaped structure and filaments was zoomed in (middle). NWT exhibited strong fluorescence signals in a minigenome assay in BSR-T7/5 cells (right). (E) The NLoop formed filaments but was not functional in a minigenome assay. The retention volume of the NLoop was ~47 ml, close to NWT (left). Negative-stain EM showed more filaments than NWT (middle). However, there was no fluorescence signal in the minigenome assay (right).
The summary and comparison of N and the derived mutants in the nucleocapsid assembly and their function in a minigenome assay.
Six N proteins including NWT, N∆C-arm∆C-tail, N∆C-tail, N∆N-arm∆C-arm∆C-tail, N∆N-arm and NLoop were subjected to gel filtration chromatography (left), negative stain EM (middle) and minigenome analyses (right). The negative stain image of NWT was cropped from Figure 1—figure supplement 1B for convenient comparation.
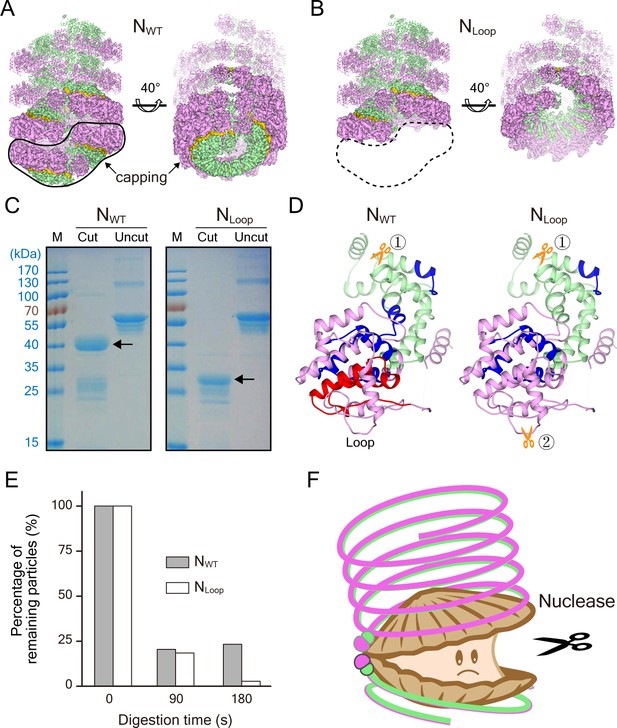
Clam-shaped nucleocapsid is resistant to elastase and RNase A.
(A) An atomic model of an NWT double-headed filament from different views shows reciprocal capping between two single-headed spirals. Colors as in Figure 1. One single-headed spiral is highlighted by the black line and labeled ‘capping’. (B) An atomic model of an NLoop single-headed filament from different views with no cap and with the 5′ end of its RNA exposed. The supposed capping spiral, marked by the dashed line, is missing from the single-headed filament. (C) SDS-PAGE gels of NWT and NLoop after elastase digestion. There was a ~40-kDa main band with some smears in the elastase-digested NWT assay (left), while elastase cut the NLoop to form a ~30-kDa band (right). (D) Mass spectrum results identified peptides drawn on the atomic structure of N, indicating one additional cutting site on NLoop (gold scissors). The common regions mapped by Mass Spectrum in NWT and NLoop are colored in blue, and the unique region checked in NWT is shown in red. Five peptides were identified from the 40-kDa band of NWT and marked on the N atomic structure, leaving the CTD loops as the cutting site. Only four peptides were identified from the 30-kDa band of NLoop and marked on N atomic structure. Given the reduced molecular weight and the missing NTD peptide, another cutting site should exist within NTD. (E) Comparison of RNase A digesting NWT and NLoop at different timepoints. Both the clam-shaped structures and the filaments are counted. The numbers of oligomers in NWT and NLoop at 0 s are normalized to 100%. At 180 s, almost 100% disassembly of nucleocapsid was seen in NLoop whereas over 25% of filaments remained in NWT. (F) A cartoon depicts the hypothetical full protection provided to the viral RNA genome by NWT via the self-capping clam-shaped structure. When the clam-shaped structure is broken, nuclease is able to access the RNA 5’ end and can digest the whole RNA strand.
Peptide mapping and RNase A digestion of NWT and NLoop.
(A) Peptides identified via Mass Spectrum are marked. Similar identified peptides, including residues 207–222, 229–237, 240–260, 353-361 in both NWT and NLoop, are colored in blue. The peculiar identified peptides 33–85 in NWT are colored in red. The gold scissors indicate the possible proteolytic sites. (B) Negative-stain EM images of RNase A digestion on NWT and NLoop. Typical micrographs of NWT and NLoop at different timepoints are presented.

C-tail may be located inside the clam-shaped structure.
(A) C-tail density is boosted by lowering the density threshold of the 3D structure without C2 symmetry. Two bulks of cone-like density (violet) emerge near the end of the docked CTD (green). Both side view (left) and top view (right) are provided for better visualization of the C-tail. (B) A typical 2D averaged class (left) shows the extra density assigned to the C-tail in the 3D structure, and this 2D-averaged class is colored in violet and yellow (right).
Videos
3D reconstruction of the clam-shaped structure and the fitting of a pseudoatomic model.
https://doi.org/10.7554/eLife.45057.007Morphing of the ring structure to form a single-turn spiral.
https://doi.org/10.7554/eLife.45057.008RNA enwrapped between the NTD and the CTD.
https://doi.org/10.7554/eLife.45057.009Double-headed spiral derived from clam-shaped structure.
https://doi.org/10.7554/eLife.45057.010Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Newcastle disease virus) | NDV N | Sangon Biotech Company | GenBank ID: HM063424.1 | Synthetic gene |
| Strain, strain background (E. coli) | BL21 (DE3) Star competent cells | ThermoFisher Scientific | C6010-03 | Cells for protein expression |
| Strain, strain background (Newcastle disease virus) | LaSota | China Veterinary Culture Collection Center | ||
| Cell line (hamster) | BSR-T7/5 | PMID: 9847328 | Gift from Zhigao Bu's lab from Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences | |
| Chemicalcompound, drug | elastase | SIGMA | E8140-1UN | |
| Chemicalcompound, drug | RNase A | Promega | A7973 | |
| Software, algorithm | RELION 1.4 | PMID: 23000701 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page | |
| Software, algorithm | RELION 2.0 | PMID: 27845625 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page | |
| Software, algorithm | UCSF Chimera | http://plato.cgl.ucsf.edu/chimera/ | RRID:SCR_004097 | |
| Software, algorithm | Coot | PMID: 20383002 | RRID:SCR_014222 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Software, algorithm | PHENIX | PMID: 22505256 | RRID:SCR_014224 | https://www.phenix-online.org/ |
| Software, algorithm | ImageJ | http://imagej.nih.gov/ij/ | RRID: SCR_003070 | |
| Other | Crystal structure of the paramyxovirus parainfluenza virus 5 nucleoprotein–RNA complex | PMID: 25831513 | PDB: 4XJN |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45057.019





