Conformational dynamics between transmembrane domains and allosteric modulation of a metabotropic glutamate receptor
Figures
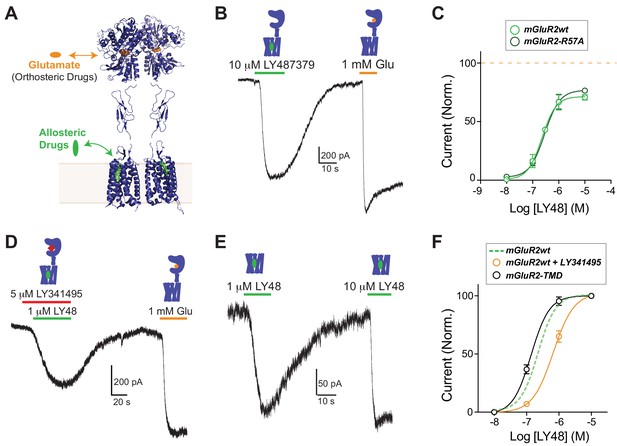
Positive allosteric modulators activate mGluR2 in a GIRK activation assay independently of ligand binding domains.
(A) Structural model of a full-length mGluR based on structures of isolated domains (mGluR2 Ligand Binding Domain in Closed-Closed/Active state with two bound glutamate molecules = PDB 5CNI; mGluR2 Cysteine-Rich Domain = PDB 5KZQ; mGluR1 Transmembrane Domain = PDB 4OR2) showing the locations of the orthosteric and allosteric binding sites. (B) Representative whole cell patch clamp recording from HEK 293T cells expressing full-length mGluR2 showing an inward GIRK current induced by the positive allosteric modulator LY483739 (LY48) that is comparable to the response to saturating glutamate. (C) LY48 GIRK activation dose response curves for wild-type SNAP-mGluR2 (EC50 = 0.23 ± 0.04 µM) and SNAP-mGluR2-R57A (EC50 = 0.29 ± 0.08 µM). Values are normalized to saturating glutamate and come from at least three cells per conditions. Error bars show s.e.m. (D–E) LY48 GIRK responses are not blocked by a saturating concentration of the competitive orthosteric antagonist LY341495 (D) or by the removal of the extracellular domain of mGluR2 (E). (F) LY48 GIRK activation dose response curves showing the apparent affinity shifts observed when LY341495 is co-applied (orange; EC50 = 0.66 ± 0.07 µM) or when the isolated mGluR2 TMD (black; EC50 = 0.14 ± 0.01 µM) is tested. Values are normalized to saturating LY48 and come from at least three cells per conditions. Error bars show s.e.m. Note: N-terminally SNAP-tagged constructs were used for all recordings.
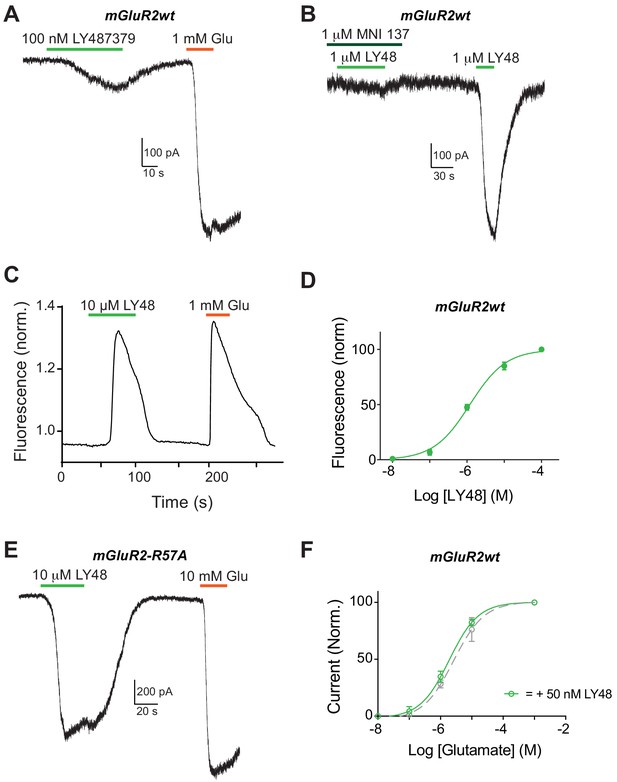
Further characterization of mGluR2 activation by LY483739.
(A–B) Representative whole cell patch clamp recordings from HEK 293T cells expressing full-length mGluR2 showing inward GIRK currents induced by the positive allosteric modulator LY483739 (LY48). Sub-saturating concentrations of LY48 produce weak activation relative to saturating glutamate (A), saturating LY48-induced currents can be blocked by the negative allosteric modulator MNI-137 (B). (C) Representative trace showing the response to LY48 and glutamate in a calcium imaging assay in HEK cells co-transfected with full-length mGluR2, GCaMP6f and Gαiq. (D) Dose response curve (EC50 = 1.2 ± 0.2 µM) for LY48 produced from calcium imaging assay. Values are normalized to saturating glutamate and at least three separate experiments contribute to each point. (E) Representative trace showing that LY48-induced currents are still seen when the low-affinity R57A mutation is introduced to the glutamate binding site (C). (F) Glutamate dose-response curves for mGluR2 in the presence (EC50 = 1.9 ± 0.2 µM) or absence (EC50 = 2.6 ± 0.5 µM) of 50 nM LY48. Values are normalized to saturating glutamate and come from at least 3 cells per conditions. Error bars show s.e.m.
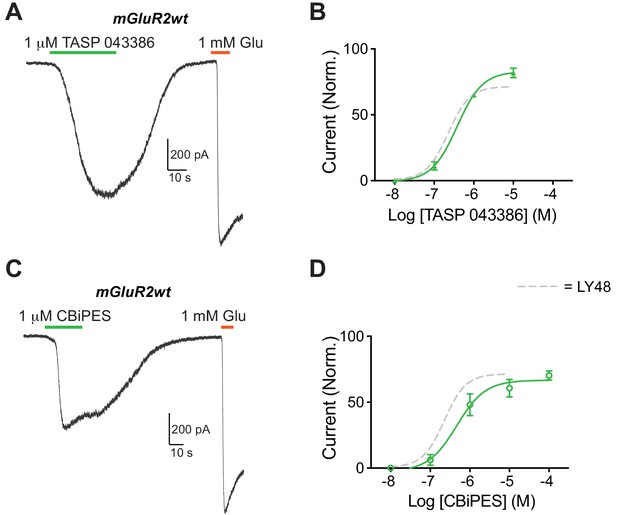
Activation of mGluR2 by the mGluR2 PAMs TASP 043386 and CBiPES.
(A–D) Representative whole cell patch clamp recordings and dose response curves for TASP (A, B; EC50 = 0.38 ± 0.11 µM) and CBiPES (C, D; EC50 = 0.46 ± 0.20 µM). Values are normalized to saturating glutamate and come from at least three cells per conditions. Error bars show s.e.m.
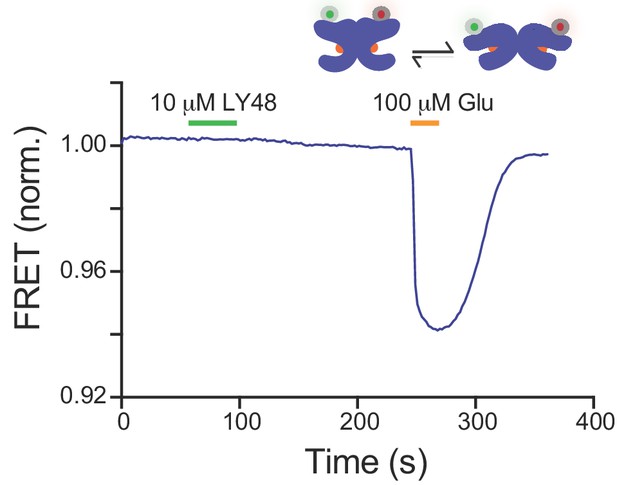
PAMs do not induce an inter-LBD FRET change in mGluR2.
Representative inter-LBD FRET measurement with SNAP-mGluR2. Application of LY48 has no effect on FRET level whereas glutamate produces a large FRET decrease due to clamshell closure and inter-LBD reorientation.
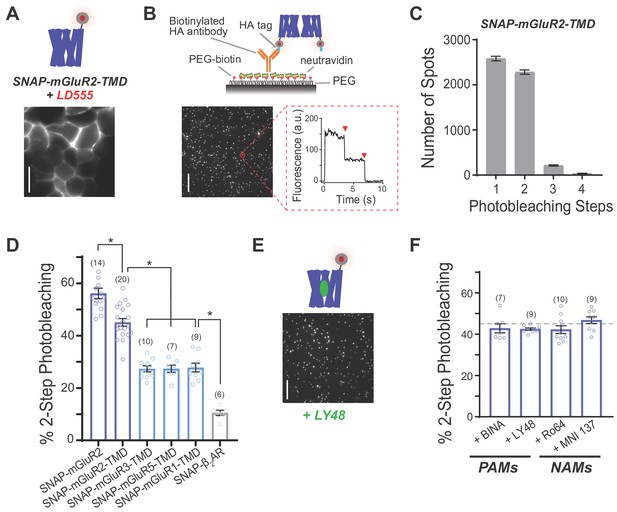
Single-molecule photobleaching analysis shows that mGluR transmembrane domains dimerize in the absence or presence of allosteric drugs.
(A) Representative image showing expression and surface labeling of SNAP-mGluR2-TMD construct in HEK 293T cells prior to lysis. Scale bar = 10 µm (B) Single-molecule pulldown of SNAP-mGluR2-TMD reveals ECD-independent dimerization of surface-targeted TMDs. Top, schematic showing the SiMPull assay using a PEG-passivated glass coverslip and a biotinylated anti-HA antibody. Bottom, representative image of single molecules with representative fluorescence time course for an individual molecule (red circle) showing two-step photobleaching (red arrows). Scale bar = 10 µm. (C) Histogram summarizing the distribution of photobleaching step number for all SNAP-mGluR2-TMD molecules tested (n = 5113 spots from 20 movies). (D) Summary bar graph showing the percentage of spots bleaching in two steps for a range of N-terminally SNAP-tagged constructs labeled with LD555. * indicates statistical significance (unpaired t tests; for SNAP-mGluR2 vs. SNAP-mGluR2-TMD, p=0.0001; for SNAP-mGluR2-TMD vs. SNAP-mGluR3-TMD, SNAP-mGluR5-TMD or SNAP-mGluR1-TMD, p=3E-10, 3E-8, and 2E-7; for SNAP-ß2AR vs. SNAP-mGluR3-TMD, SNAP-mGluR5-TMD or SNAP-mGluR1-TMD, p=4E-8, 1E-6, and 1E-6). The number of movies analyzed for each condition is shown in parentheses. Error bars show s.e.m. (E) Representative image showing immobilized SNAP-mGluR2-TMD molecules treated with LY48. Scale bar = 10 µm. (F) Summary bar graph showing the lack of an effect of application of PAMs (10 µM LY48 or 1 µM BINA) or NAMs (10 µM Ro 64–5229 or 1 µM MNI 137) on SNAP-mGluR2-TMD stoichiometry. Dotted gray line shows the % two-step bleaching observed for un-liganded SNAP-mGluR2-TMD. The difference between conditions, including WT, were not significant (ANOVA, p=0.2). The number of movies analyzed for each condition is shown in parentheses. Error bars show s.e.m.
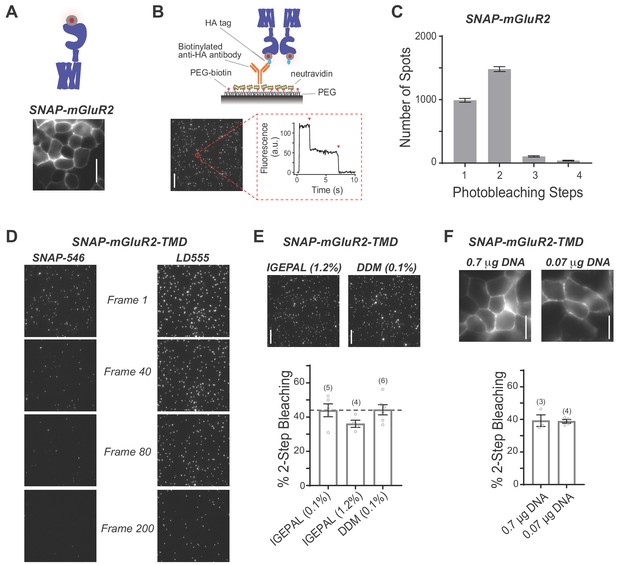
Further analysis of single-molecule pulldown of mGluR2 and mGluR2-TMD.
(A) Representative image showing expression and surface labeling of SNAP-mGluR2 construct in HEK 293T cells prior to lysis. Scale bar = 10 µm. (B) Single-molecule pulldown of SNAP-mGluR2 confirms dimerization of surface-targeted full-length receptors. Top, schematic showing the SiMPull assay using a PEG-passivated glass coverslip and a biotinylated anti-HA antibody. Bottom, representative image of single molecules with representative fluorescence time course for an individual molecule (red circle) showing two-step photobleaching (red arrows). Scale bar = 10 µm. (C) Histogram summarizing the distribution of photobleaching step number for all SNAP-mGluR2 molecules tested (n = 2618 spots from 14 movies). (D) LD555 significantly improves fluorophore lifetime compared to commercially available SNAP Alexa- 546. Identical laser intensities and imaging speeds (continuous 20 Hz imaging) were used for samples labeled with either fluorophore. (E) Test of different detergent conditions for SiMPull with SNAP-mGluR2-TMD, showing that identical results are observed in IGEPAL and DDM at high and low concentrations. The number of movies analyzed for each condition is shown in parentheses. Error bars show s.e.m. (F) Transfecting HEK293T cells with a 10-fold decrease in SNAP-mGluR2-TMD DNA does not alter photobleaching step distribution despite a modest decrease in receptor expression. The number of movies analyzed for each condition is shown in parentheses. Error bars show s.e.m. All scale bars = 10 µm.
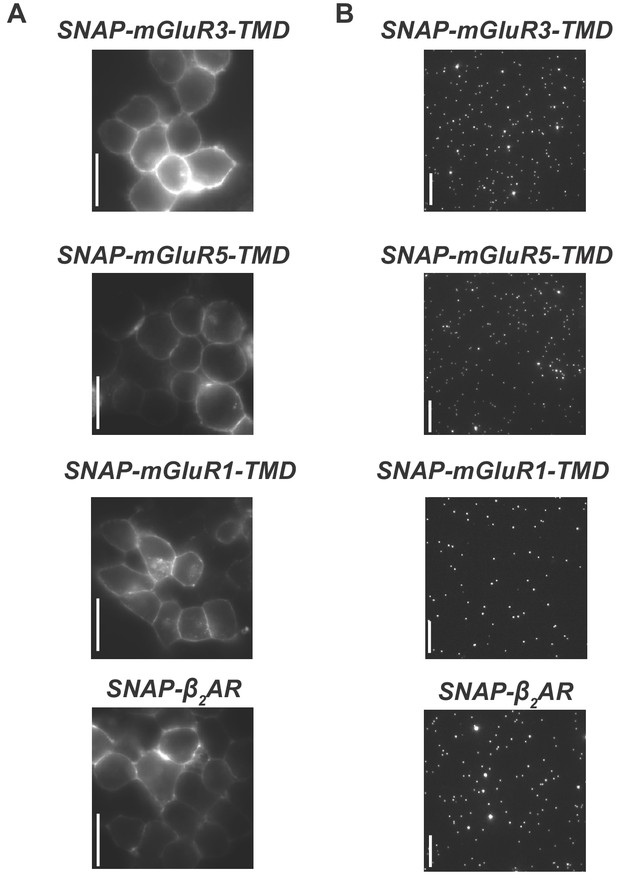
Comparative analysis of GPCR TMD stoichiometry: mGluR2, mGluR3, mGluR5 and ß2AR Representative HEK 293T expression.
(A) and single molecule images (B) for SNAP-mGluR3-TMD, SNAP-mGluR5-TMD, SNAP-mGluR1-TMD and SNAP-ß2AR. All scale bars = 10 µm.
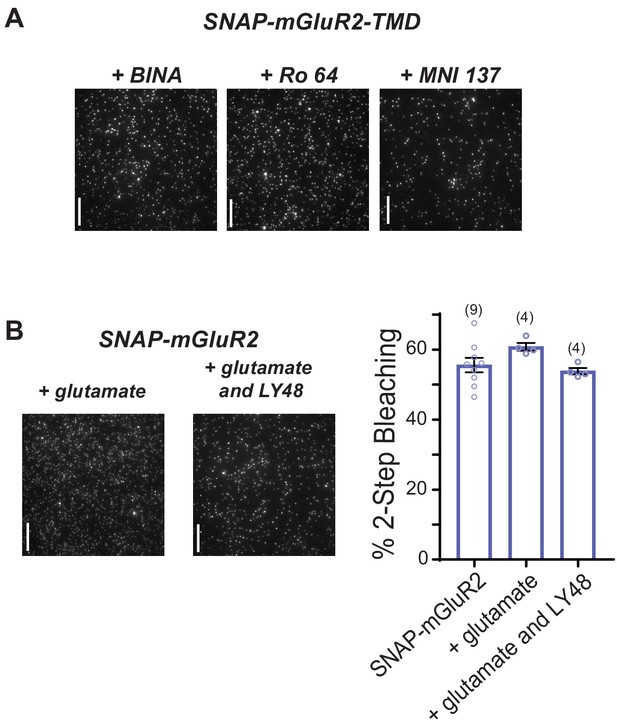
Further analysis of the lack of effect of ligands on mGluR2 stoichiometry.
(A) Representative images of immobilized SNAP-mGluR2-TMD molecules treated with either 1 µM BINA (PAM), 10 µM Ro 64 (NAM) or 1 µM MNI 137 (NAM). (B) Left, representative images of immobilized SNAP-mGluR2 molecules treated with either 1 mM glutamate or 1 mM Glutamate and 10 µM LY48. Right, summary bar graph showing that drug treatment does not alter % two-step bleaching for SNAP-mGluR2. Error bars show s.e.m. All scale bars = 10 µm.
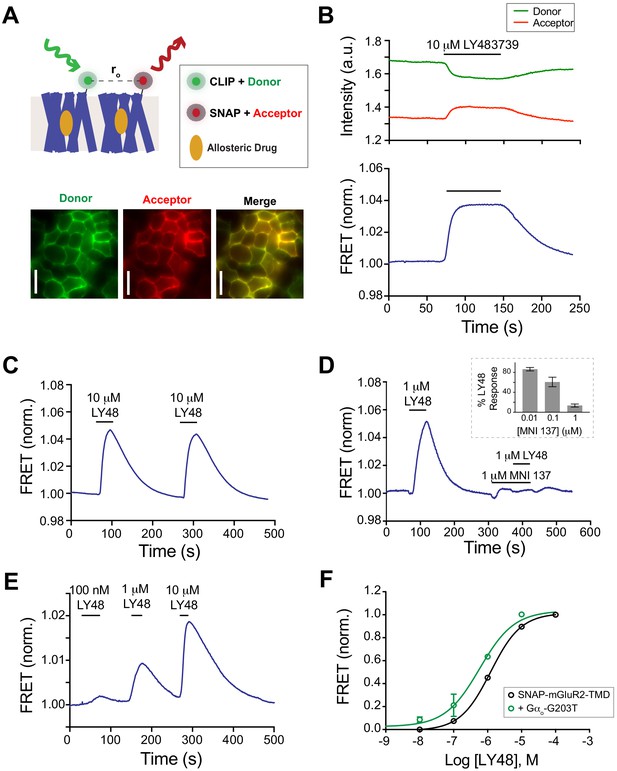
An inter-TMD FRET assay reveals LBD-independent reorientation in response to positive allosteric modulators.
(A) Top, schematic showing SNAP- and CLIP-tagged mGluR2-TMD constructs labeled with donor and acceptor fluorophores. Bottom, images showing donor and acceptor channels following donor excitation with a 561 nm laser. Scale bars = 40 μm. (B) Representative time course showing donor and acceptor fluorescence intensity (top) during LY48 application. Baseline-normalized FRET is shown in the bottom trace, revealing a large, reversible increase in response to LY48 application. (C–E) Representative traces showing that LY48-induced inter-TMD FRET increase is repeatable (C), blocked by the NAM MNI 137 (D), and dose-dependent (E). The inset to (D) shows the extent of block of a 10 μM LY48 response by different concentrations of MNI 137. (F) Dose–response curve for LY48-induced FRET increase for SNAP-mGluR2-TMD (EC50 = 1.2 ± 0.1 µM), WT-SNAP-mGluR2 + dominant negative G protein (EC50 = 0.6 ± 0.04 µM). The dose–response curves were significantly different (two-way ANOVA, p=0.002). Values are normalized to saturating LY48 and come from at least three separate experiments per conditions. Error bars show s.e.m.
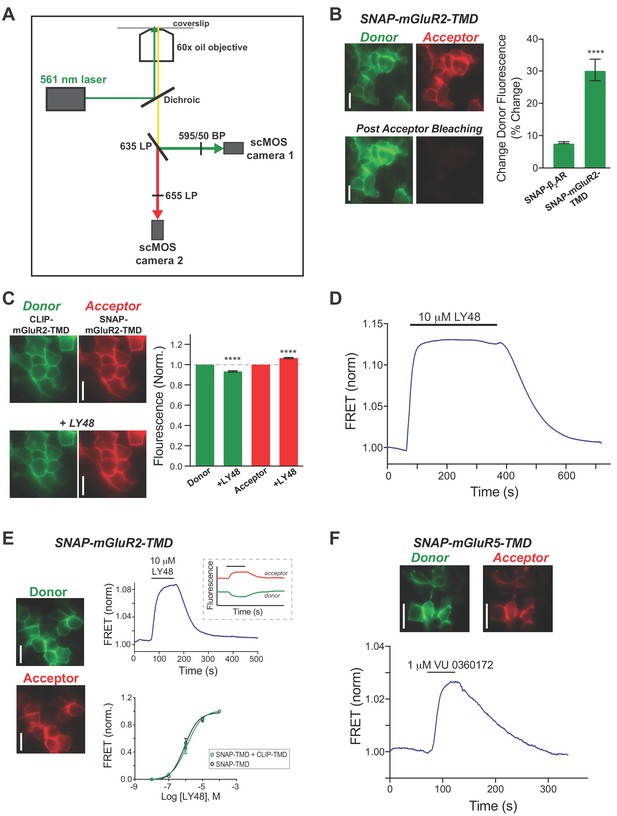
Further characterization of an inter-TMD mGluR FRET Assay.
(A) Configuration of inverted microscope used for ensemble FRET imaging. Briefly, 560 nm laser illumination is used for excitation and donor and acceptor emission is separated with a dichroic in front of two separate, aligned cameras. See Materials and methods for more details. (B) Left, representative donor and acceptor images of HEK 293T cells before and after acceptor channel bleaching for SNAP-mGluR2-TMD. Right, summary bar graph showing a significantly higher donor recovery for SNAP-mGluR2-TMD compared to the prototypical class A GPCR, SNAP-ß2AR (unpaired t test, p=0.003). Error bars show s.e.m. (C) Left, representative images of HEK 293T cells expressing SNAP-mGluR2-TMD before and after application of 10 μM LY48. Right, summary bar graph showing an increase in acceptor (paired t test, p=0.00004) and decrease (paired t test, p=0.00005) in donor fluorescence following LY48 application. Error bars show s.e.m. (D) Representative inter-TMD FRET trace showing that the response to LY48 is sustained and non-desensitizing during a 5-min period. (E) Summary of results showing the SNAP-mGluR2-TMD expression and labeling with donor (BG-LD-555) and acceptor (BG-LD-655) shows identical FRET responses to co-expression of SNAP-mGluR2-TMD and CLIP-mGluR2-TMD. Left, representative images showing donor- and acceptor-labeled SNAP-mGluR2-TMD-expressing HEK 293T cells. Top right, representative FRET trace showing an increase in inter-TMD FRET in response to LY48 application. The inset shows the donor (green) and acceptor (red) channels separately. Bottom right, dose–response curve showing identical curves for SNAP-mGluR2-TMD labeled with donor and acceptor (EC50 = 1.2 ± 0.1 µM) versus co-expression of SNAP- and CLIP-mGluR2-TMD (EC50 = 0.9 ± 0.1 µM). Values are normalized to saturating LY48 and come from at least three separate experiments per conditions. Error bars show s.e.m. (F) Representative images (top) and inter-TMD FRET trace (bottom) for SNAP-mGluR5-TMD in response to the mGluR5 PAM VU 0360172 showing that the LBD-independent rearrangement of TMDs is observed across the mGluR family. All scale bars = 20 µm.
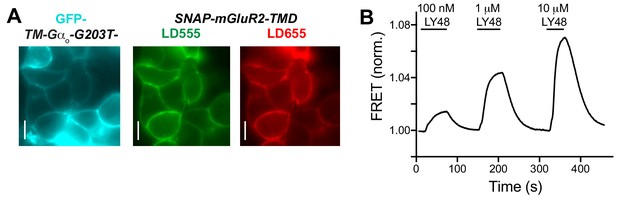
Further analysis of G protein effects on inter-TMD rearrangement.
(A) Representative images of HEK 293T cells expression GFP-TM-GαoA-G203T (blue) and SNAP-mGluR2-TMD labeled with donor (green) and acceptor (red). Scale bars = 20 µm. (B) Representative inter-TMD FRET trace showing dose-dependent LY48-induced increases in FRET.
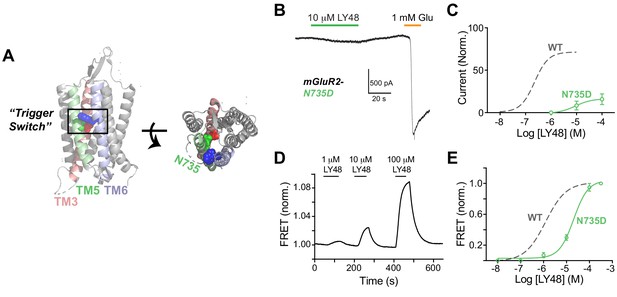
A role for a TMD ‘Trigger Switch’ in allosteric agonism and inter-TMD re-arrangement.
(A) mGluR1-TMD crystal structure showing the location of amino acids in TM3, 5 and 6 proposed to form the PAM ‘trigger switch’ including N735 in mGluR2 (red). (B) Representative HEK 293T cell patch recording showing a weak LY48-induced GIRK response relative to saturating glutamate in cells expressing full-length mGluR2. (C) Dose–response curve showing a large decrease in GIRK activation by LY48 for mGluR2-N735D. Values are normalized to saturating glutamate and come from at least three separate experiments per conditions. Error bars show s.e.m. (D) Representative inter-TMD FRET trace showing the response to LY48 for SNAP-mGluR2-TMD-N735D. (E) LY48 dose–response curves showing a large right-shift for SNAP-mGluR2-TMD-N735D (EC50 = 20.5 ± 1.4 µM) relative to wildtype (EC50 = 1.2 ± 0.1 µM). Values are normalized to saturating LY48 and come from at least three separate experiments per conditions. Error bars show s.e.m.
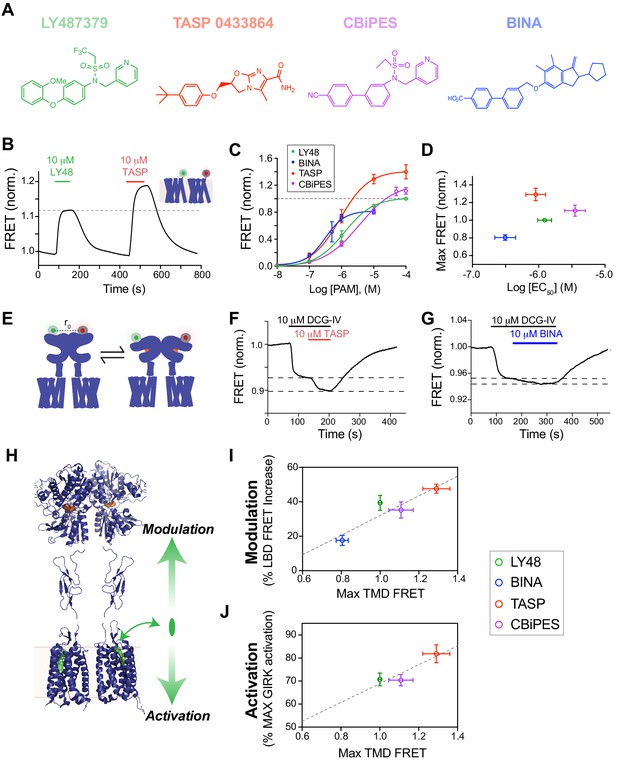
Structurally distinct PAMs increase inter-TMD FRET with variable potency and efficacy which is correlated to the strength of both allosteric activation and modulation.
(A) Chemical structures of mGluR2 PAMs. (B) Representative trace showing inter-TMD FRET responses to LY48 and TASP. At saturating concentrations, TASP shows a larger FRET response than LY48. (C) Dose-response results for all four PAMs tested in the inter-TMD FRET assay, normalized to saturating LY48. (LY48 EC50 = 1.2 ± 0.1 µM, TASP EC50 = 0.9 ± 0.1 µM, CBiPES EC50 = 3.5 ± 0.2 µM, BINA EC50 = 3.1 ± 0.2 µM). Values are normalized to saturating LY48 and come from at least three separate experiments per condition. Error bars show s.e.m. (D) Summary of inter-TMD efficacy (i.e. max amplitude) and apparent affinity (i.e. EC50) for all PAMs tested. Note the lack of correlation between relative efficacy and relative affinity. (E) Schematic of inter-LBD FRET assay where SNAP-tagged full-length mGluR2 constructs are labeled with donor and acceptor fluorophores and an orthosteric agonist is applied, leading to a decrease in FRET. (F–G) Representative inter-LBD FRET traces showing variability of the magnitude of PAM potentiation of the response to a saturating concentration of an mGluR2 agonist (DCG-IV). TASP (F) produces a larger increase in the FRET response than BINA (G). (H) Schematic showing that PAMs are able to exert their functional effects on mGluR2 in two different directions, either through alteration of the response to orthosteric agonists (‘modulation’) or through direct control of receptor signaling (‘activation’). Error bars show s.e.m. (I–J) Plots showing correlation between inter-TMD FRET efficacy and either allosteric modulation (I; R2 = 0.84) or allosteric activation (J; R2 = 0.84).
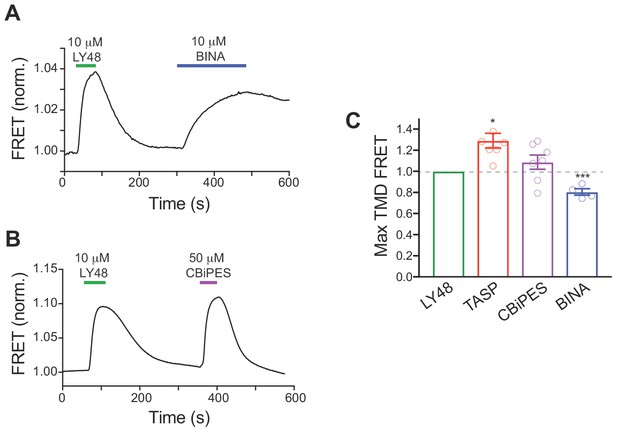
Further comparative analysis of PAM-mediated inter-TMD FRET changes and GIRK activation.
(A–B) Representative traces showing the relative inter-TMD FRET changes observed for saturating BINA (A) and CBiPES (B) relative to LY48. (C) Summary bar graphs showing maximum inter-TMD FRET for all four PAMs tested. Values are normalized to saturating LY48. * indicates statistical significance relative to LY48 (unpaired t tests: p=0.003 for TASP and p=0.00005 for BINA). Values come from at least three separate experiments per conditions. Error bars show s.e.m.
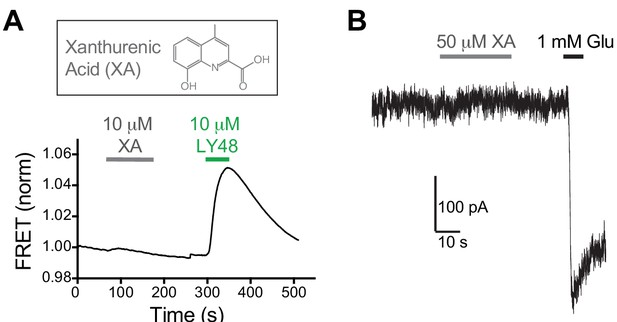
Xanthurenic acid does not serve as an allosteric agonist of mGluR2 representative inter-TMD FRET.
(A) and whole cell patch recording of full-length mGluR2 (B) showing no effect of Xanthurenic acid. Identical results were seen in >3 independent experiments for both FRET and patch measurements.
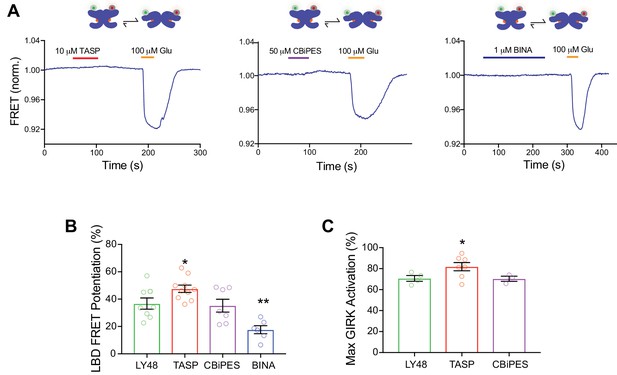
Comparison of allosteric activation and modulation efficacies of mGluR2 PAMs.
(A) Representative inter-LBD FRET traces showing no FRET response for TASP , CBiPES or BINA . Identical results were seen in >3 independent experiments. (B) Summary bar graph showing the increase in LBD FRET response in the presence of 10 µM DCG-IV for all four mGluR2 PAMs at saturating concentrations (10 µM for LY48, BINA and TASP; 50 µM for CBiPES). * indicates statistical significance relative to LY48 (unpaired t tests: p=0.004 for BINA and p=0.03 for TASP). Values come from at least three separate experiments per conditions. Error bars show s.e.m. (C) Summary bar graph showing the maximum GIRK response relative to 1 mM glutamate for each mGluR2 PAM. * indicates statistical significance relative to LY48 (unpaired t test, p=0.03). Values come from at least three separate experiments per conditions. Error bars show s.e.m.
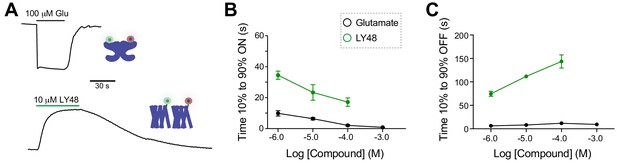
Comparative analysis of kinetics of glutamate-induced LBD FRET changes and LY48-induced TMD FRET changes.
(A) Representative inter-LBD (top) and inter-TMD (bottom) FRET traces showing the timing of responses to saturating glutamate and LY48, respectively. (B) Summary graph showing the dose-dependence of the ON kinetics of ligand-induced inter-TMD or inter-LBD FRET changes. * indicates statistical significance between glutamate and LY48 responses at a given concentration (unpaired t tests: for 1 µM, p=0.02; for 10 µM, p=0.04; for 100 µM, p=0.03). (C) Summary graph showing the dose-dependence of the OFF kinetics of ligand-induced inter-TMD or inter-LBD FRET changes. * indicates statistical significance between glutamate and LY48 responses at a given concentration (unpaired t tests: for 1 µM p=0.0003; for 10 µM, p=0.0007 µM; for 100 µM p=0.001). Error bars show s.e.m. Values come from at least three separate measurements per conditions.
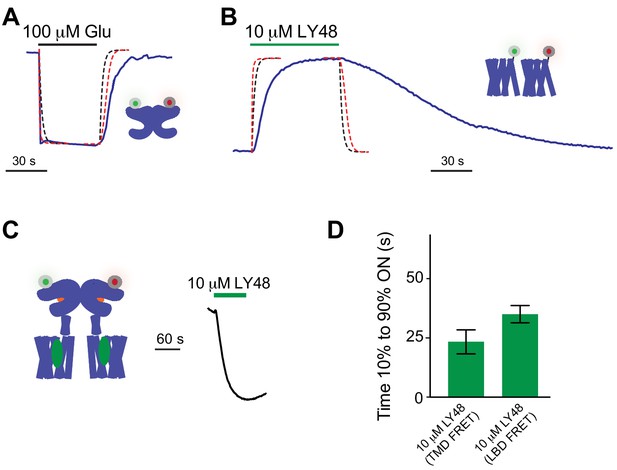
Further analysis of glutamate-induced inter-LBD kinetics and LY48-induced inter-TMD kinetics.
(A–B) Calculated traces using a model with simple ligand binding and un-binding to an accessible site showing the expected time course of changes in the bath concentration of drug (black dotted line) and the expected response for a concentration ten times the EC50 (red dotted line; values calculated using a Hill equation with n = 1) for the experimental system (25 mL/minute flow with a 0.5 mL chamber and uniform mixing) compared to raw data (blue line) for glutamate-induced LBD FRET changes (A) or LY48-induced TMD FRET changes (B). (C) Representative trace showing the inter-LBD FRET response to LY48 in the presence of 10 µM DCG-IV. (D) Summary bar graph showing similar ON kinetics of LY48 response in both inter-TMD and inter-LBD FRET assays.
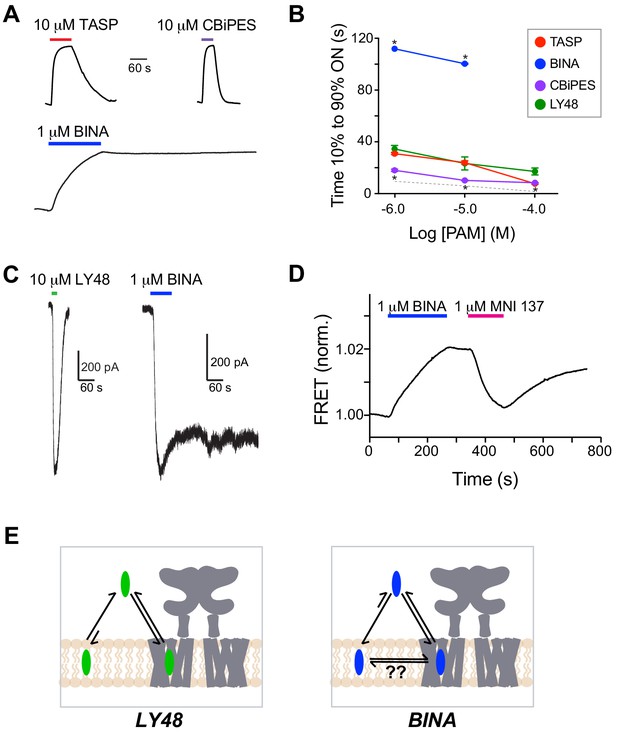
Different mGluR2 PAMs alter inter-TMD FRET and receptor activation with distinct kinetics and reversibility.
(A) Representative inter-TMD FRET traces for TASP, CBiPES and BINA showing slower onset and lack of reversibility for BINA. (B) Summary of dose-dependent ON kinetics for all four mGluR2 PAMs tested BINA was significantly slower (unpaired T-test; 1 μM: p=0.000002, 10 μM: p=0.007) and CBiPES was significantly faster (unpaired T-test; 1 μM: p=0.04, 10 μM: p=0.03, 100 μM: p=0.02) than LY48. At least three separate measurements were made for each condition. Error bars show s.e.m. The dotted gray line shows the values obtained for glutamate in the inter-LBD FRET response. (C) Whole cell patch clamp recordings showing reversibility of LY48 agonism and irreversibility of BINA agonism of full-length mGluR2. (D) Representative inter-TMD FRET trace showing the effect of application of an mGluR2 NAM (MNI 137) following BINA application. Notably, MNI 137 reverses the BINA-induced FRET increase but following MNI 137 washout, the FRET level increases again. (E) Schematics showing working model of PAM binding and membrane interaction for LY48 versus BINA. Unlike other PAMs, BINA may partition directly into the lipid bilayer from where it can access the allosteric binding site on mGluR2 either directly within the bilayer or following exit on the extracellular side.
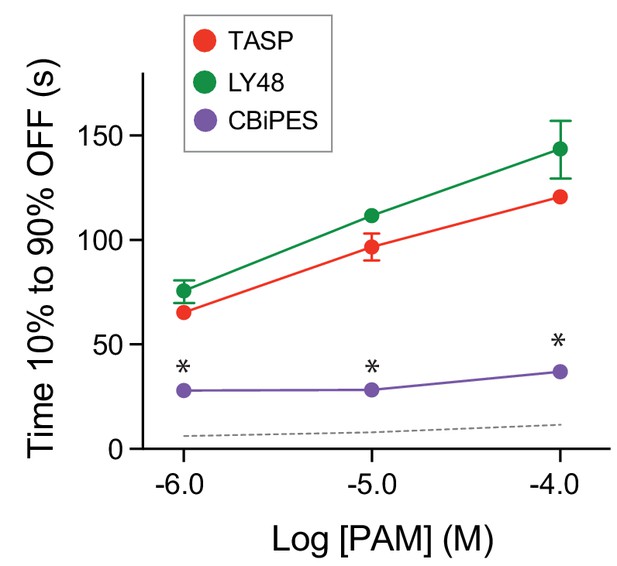
OFF kinetics of PAM-induced conformational changes of mGluR2 .
Summary graph showing the dose-dependence of the OFF kinetics of ligand-induced inter-TMD FRET changes. * indicates statistical significance of CBiPES responses relative to LY48 responses at a given concentration (unpaired t tests; 1 μM: p=0.00009, 10 μM: p=0.0003, 100 μM: p=0.002). Values come from at least three separate measurements per conditions. Error bars show s.e.m. The dotted gray line shows the values obtained for glutamate in the inter-LBD FRET response.
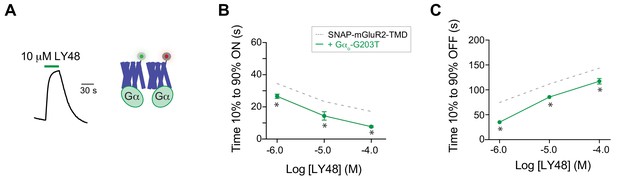
Analysis of the effects of G protein on LY48-induced inter-TMD FRET kinetics.
(A) Representative inter-TMD FRET responses for LY48 in the presence of a dominant negative Gα protein. (B) Summary graph showing the dose-dependence of the ON kinetics of LY48-induced inter-TMD FRET changes. * indicates statistical significance relative to responses without dominant negative Gα at a given concentration (unpaired t tests: for 1 µM, p=0.04; for 10 µM, p=0.03; for 100 µM, p=0.02). Values come from at least three separate measurements per conditions. (C) Summary graph showing the dose-dependence of the OFF kinetics of LY48-induced inter-TMD FRET changes. * indicates statistical significance relative to responses without dominant negative Gα at a given concentration (unpaired t tests: for 1 µM, p=0.002; for 10 µM, p=0.002; for 100 µM, p=0.05). Values come from at least three separate measurements per conditions. Error bars show s.e.m.
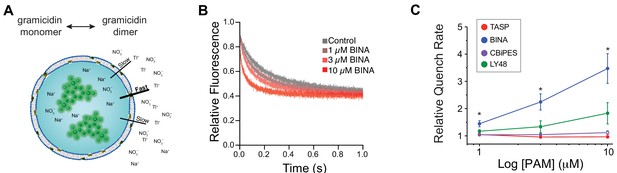
Analysis of the effects of G protein on LY48-induced inter-TMD FRET kinetics.
(A) Schematic describing liposome-based membrane interaction assay using gramicidin as a sensor of membrane properties. The gramicidin monomer↔dimer equilibrium varies with changes in lipid bilayer properties, such as thickness and elasticity. Shifts toward the right will increase the number of conducting dimers, which can be detected by measuring the rate by which the gramicidin channel permeant heavy ion quencher thallium (Tl+) quenches intravesicular fluorophores like 8-aminonaphthalene-1,3,6-trisulfonate (ANTS). (B) Representative fluorescence quench traces recorded by mixing ANTS-loaded LUVs that have been equilibrated with BINA at the indicated concentrations with a Tl+-containing solution in a stopped-flow spectrofluorometer. (C) Dose–response curves for all PAMs showing relative effects on quenching rate in fluorescence membrane interaction assay. * indicates statistical significance (unpaired t-test with Holm-Sidak correction, p=0.2 for 1 µM BINA, p=0.02 for 3 µM BINA and p=0.03 for 10 µM BINA). Values come from at least three separate measurements per conditions. Error bars show s.e.m.
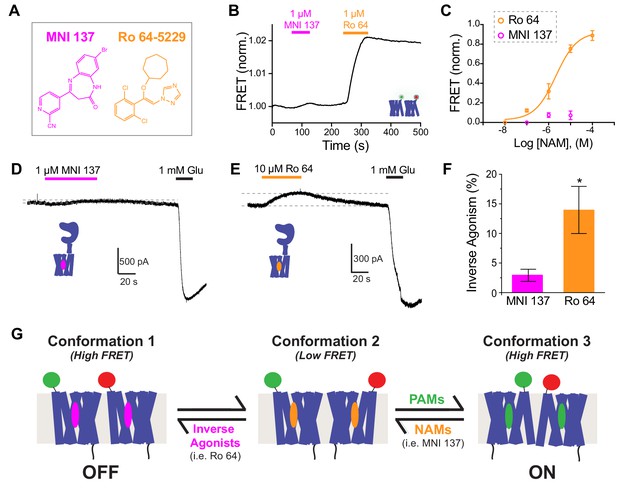
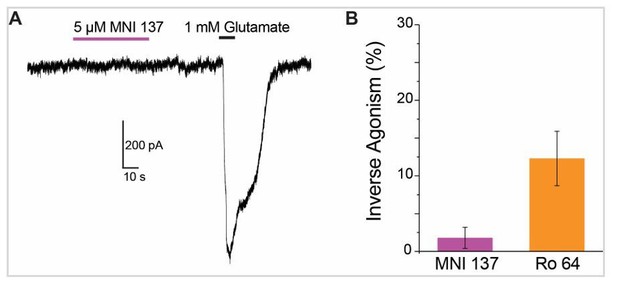
Inverse agonism or neutral antagonism of NAMs is associated with the presence or absence of an inter-TMD FRET Response.
(A) Chemical structures of the mGluR2 NAMs, MNI 137 and Ro 64–5229. (B) Representative inter-TMD FRET trace showing the lack of a response for MNI 137, but a large FRET increase in response to Ro 64 application. (C) Inter-TMD FRET dose–response curves showing a large dose-dependent response for Ro 64 with an EC50 of 2.1 ± 0.2 μM. Values are normalized to saturating LY48 and come from at least three separate experiments per conditions. Error bars show s.e.m. (D–F) Functional evidence for neutral antagonism of MNI 137 and inverse agonism of Ro 64. Representative current traces show no effect on baseline levels for MNI 137 (G), but a clear outward current for Ro 64 (H). Results are summarized in (F). Error bars show s.e.m. * indicates statistical significant (unpaired t-test, p=0.04). (J) Working three-state model accommodating a FRET increase for application of either PAMs or inverse agonists. A combination of distinct inter-TMD interfaces and differences in intra-TMD conformation likely underlie the different FRET values.
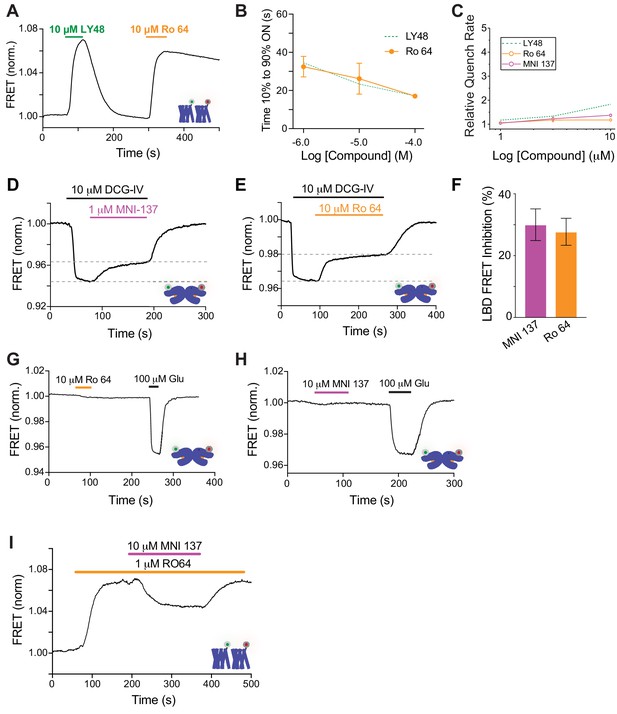
Further characterization of functional and conformational effects of mGluR2 NAMs.
(A) Representative inter-TMD FRET trace showing FRET increases in response to both LY48 and Ro 64. Note: both responses show similar ON kinetics but the OFF rate is much slower for Ro 64. (B) Summary graph showing dose-dependence of ON kinetics of Ro 64 inter-TMD FRET changes. Values come from at least three separate measurements per conditions. Error bars show s.e.m. (C) Dose-response curves for NAMs showing relative effects on quenching rate in fluorescence membrane interaction assay. Values come from at least three separate measurements per conditions. Error bars show s.e.m. (D–E) Representative inter-LBD FRET traces showing the effects of mGluR2 NAMs on the FRET decrease induced by the agonist DCG-IV. (F) Summary bar graph showing similar inter-LBD FRET changes for saturating concentrations of MNI 137 and Ro 64. Values come from at least three separate measurements per conditions. Error bars show s.e.m. (G–H) Representative inter-LBD FRET traces showing no direct effect of mGluR2 NAMs on inter-LBD conformation. (I) Representative inter-TMD FRET trace showing apparent antagonism of the Ro 64 FRET response by MNI 137.
Tables
| Reagent type | Designation | Source | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | HEK 293T | ATCC | ATCC Cat# CRL-3216, RRID:CVCL_0063 | |
| Transfectedconstruct (Rattus norvegicus) | SNAP-mGluR2 | Doumazane et al., 2011 | ||
| Transfected construct (Rattus norvegicus) | SNAP-mGluR2-TMD | Doumazane et al., 2011 (modified) | Following SNAP tag, mGluR2 truncatd to position Q558. | |
| Transfected construct (Rattus norvegicus) | SNAP-mGluR3-TMD | Doumazane et al., 2011 (modified) | Following SNAP tag, mGluR3 truncatd to position E567. | |
| Transfected construct (H. sapiens) | SNAP-mGluR5-TMD | Doumazane et al., 2011 (modified) | Following SNAP tag, mGluR5 truncatd to position V570. | |
| Transfected construct (Rattus norvegicus) | SNAP-mGluR1-TMD | Doumazane et al., 2011 (modified) | Following SNAP tag, mGluR1 truncatd to position V583. | |
| Transfected construct (Rattus norvegicus) | SNAP-mGluR2-R57A | Doumazane et al., 2011 (modified) | R at position 57 replaced with A. | |
| Transfected construct (Rattus norvegicus) | EGFP-TM-GαoA-G203T | Lober et al., 2006 (modified) | G at position 203 replaced with T. | |
| Transfected construct (Rattus norvegicus) | CLIP-mGluR2-TMD | Doumazane et al., 2011 (modified) | Following CLIP tag, mGluR2 truncatd to position Q558. | |
| Transfected construct (Rattus norvegicus) | CLIP-mGluR2 | Doumazane et al., 2011 | ||
| Transfected construct (Rattus norvegicus) | SNAP-mGluR2-TMD-N735D | Doumazane et al., 2011 (modified) | Following SNAP tag, mGluR2 truncatd to position Q558. N at position 735 replaced with D | |
| Transfected construct (H. sapiens) | SNAP-B2AR | modified from addgene plasmid pSNAPf-ADRβ2 | Addgene Plasmid #101123 | |
| Transfected construct (H. sapiens) | GIRK1-F137S homotetramerization mutant | |||
| Transfected construct (synthetic) | tdTomato | Addgene | Addgene Plasmid #30530 | |
| Transfected construct | Gαiq3 | Concept from Conklin et al., 1993 | Conklin et al., 1993 | |
| Transfected construct | GCaMP6f | Addgene | Addgene Plasmid #73564 | |
| Antibody | Rabbit polyclonal to HA tag (Biotin) | Abcam | ab26228, RRID:AB_449023 | 10–20 nM (1:500 dilution) |
| Chemical compound, drug | NeutrAvidin | ThermoFisher | Cat # 31000 | |
| Chemical compound, drug | mPEG | Laysan Bio | Item# BIO-PEG-SVA-5K-100MG andMPEG-SVA-5K-1g | |
| Chemical compound, drug | biotinylated mPEG | Laysan Bio | Item# BIO-PEG-SVA-5K-100MG and MPEG-SVA-5K-1g | |
| Chemical compound, drug | benzylguanine (BG)-LD555 | Scott Blanchard lab (synthesized in house) | Synthesized in-house | |
| Chemical compound, drug | benzylguanine (BG)-LD655 | Scott Blanchard lab (synthesized in house) | Synthesized in-house | |
| Chemical compound, drug | IGEPAL | Sigma Aldrich | I8896-50 | |
| Chemical compound, drug | DDM | Sigma Aldrich | 850520P | |
| Chemical compound, drug | SNAP Surface Alexa-546 | New England Biolabs | Cat # S9132S | |
| Chemical compound, drug | CLIP Surface Alexa-547 | New England Biolabs | Cat # S9233S | |
| Chemical compound, drug | DMEM | Thermo Fisher Scientific | Cat #11995073 | |
| Chemical compound, drug | Fetal Bovine Serum | Thermo Fisher Scientific | Cat #10437028 | |
| Chemical compound, drug | Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat # L3000015 | |
| Chemical compound, drug | Poly-L-lysine hydrobromide | Sigma Aldrich | P2636 | |
| Chemical compound, drug | Glutamate | Sigma Aldrich | Cat # 6106-04-3 | |
| Chemical compound, drug | Xanthurenic acid | Tocris | Cat # 4120 | |
| Chemical compound, drug | LY487379 | Tocris | Cat # 3283 | |
| Chemical compound, drug | BINA | Tocris | Cat # 4048 | |
| Chemical compound, drug | Ro 64–5229 | Tocris | Cat # 2913 | |
| Chemical compound, drug | MNI 137 | Tocris | Cat # 4388 | |
| Chemical compound, drug | TASP 0433864 | Tocris | Cat # 5362 | |
| Chemical compound, drug | VU0360172 | Tocris | Cat # 4323 | |
| Chemical compound, drug | LY341459 | Tocris | Cat # 1209 | |
| Chemical compound, drug | 22:1 phosphotidylcholine | Avanti Polar Lipids | Cat #850398C | |
| Chemical compound, drug | Aminonaphthalene trisulfonic acid | Thermo Fisher | Cat # A350 | |
| Chemical compound, drug | Thalium nitrate | Sigma Aldrich | Cat # 309230 | |
| Chemical compound, drug | Gramicidin A | Sigma Aldrich | Cat # 50845 | |
| Software, algorithm | LabVIEW | http://www.ni.com/en-us/shop/labview/labview-details.html | RRID:SCR_014325 | |
| Software, algorithm | ImageJ | http://imagej.nih.gov/ij/ | RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism | https://graphpad.com | RRID:SCR_002798 | |
| Software, algorithm | ClampX | https://www.moleculardevices.com/ | RRID:SCR_011323 | |
| Software, algorithm | Clampfit | https://www.moleculardevices.com/ | RRID:SCR_011323 | |
| Software, algorithm | Microsoft Excel | https://products.office.com/en-us/excel | RRID:SCR_016137 | |
| Software, algorithm | Origin | https://www.originlab.com/ | RRID:SCR_002815 | |
| Software, algorithm | Adobe Illustrator | https://www.adobe.com/ | RRID:SCR_010279 | |
| Software, algorithm | Olympus cellSens | www.olympus-lifescience.com/cellsens | RRID:SCR_016238 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45116.026