'Palaeoshellomics’ reveals the use of freshwater mother-of-pearl in prehistory
Figures

The double-buttons analysed in this study.
https://doi.org/10.7554/eLife.45644.003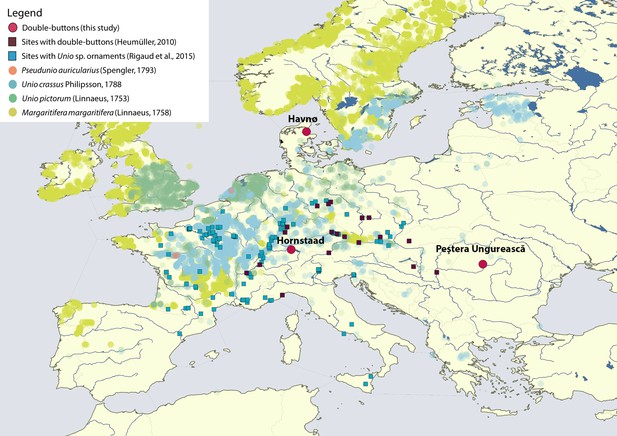
Map displaying the location of Havnø, Hornstaad-Hörnle IA and Peştera Ungurească, together with other archaeological sites from which double-buttons (Heumüller, 2012) and a variety of ornaments made with Unio sp. shells (Rigaud et al., 2015) have been reported.
https://doi.org/10.7554/eLife.45644.004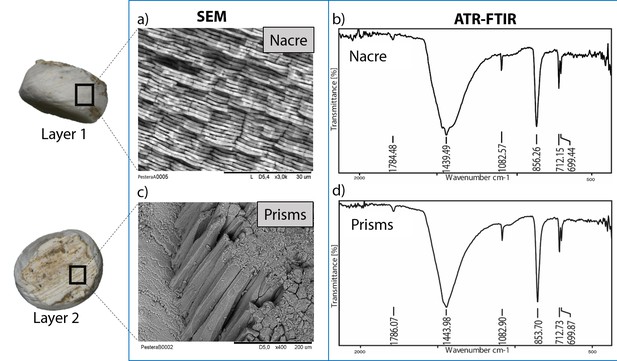
Microstructure (SEM) and mineralogy (FTIR-ATR) of double-buttons, showing shiny nacreous (a) and matte prismatic (c) layers, both aragonitic (b, d).
https://doi.org/10.7554/eLife.45644.006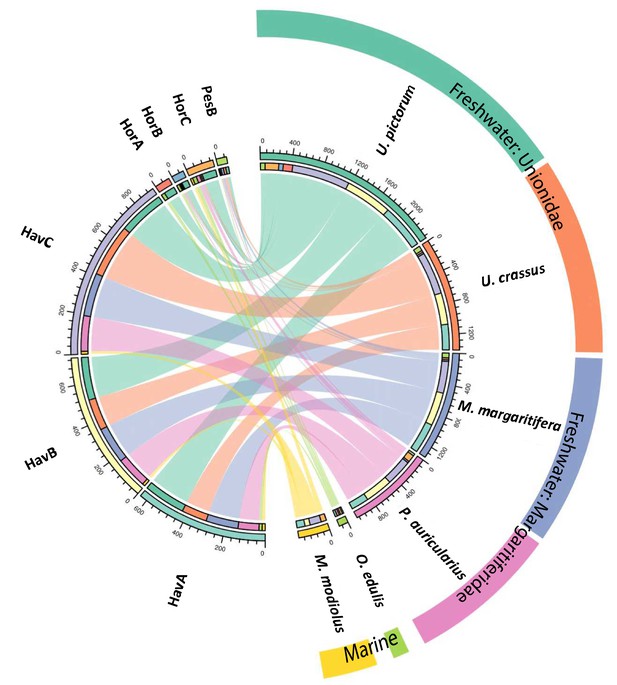
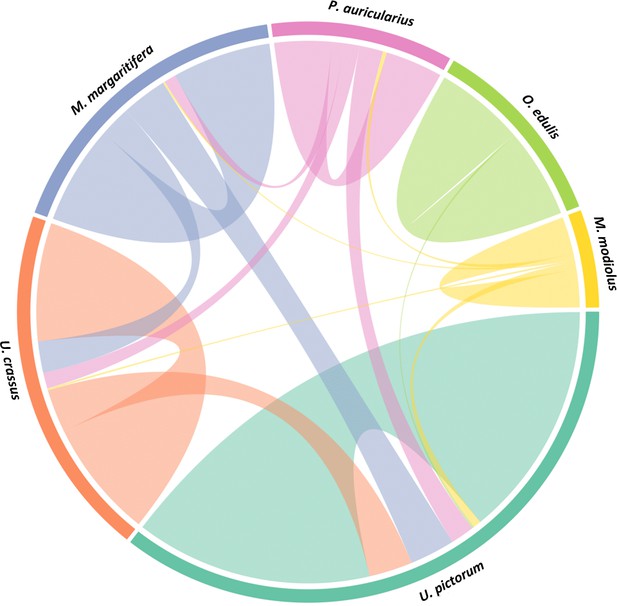
Circular diagram representing the similarity between the proteomes of seven double-buttons (left) and six mollusc shell taxa (right).
https://doi.org/10.7554/eLife.45644.009-
Figure 4—source data 1
R code and data files for Figure 4.
- https://doi.org/10.7554/eLife.45644.010
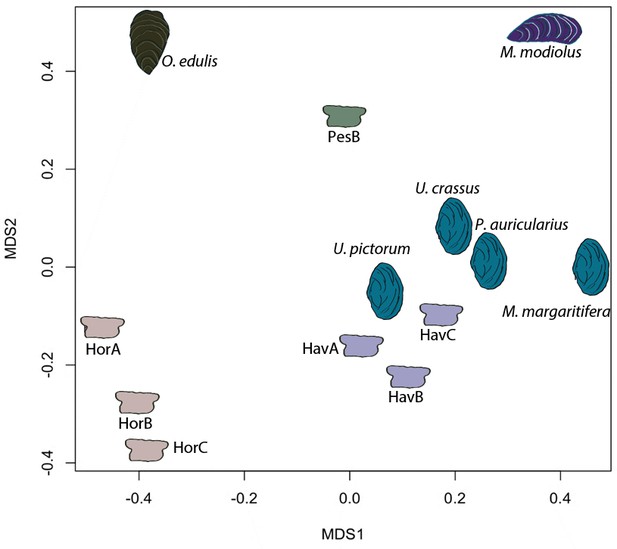
Proteome comparison based on peptide sequence similarity, represented by multi-dimensional scaling (MDS).
https://doi.org/10.7554/eLife.45644.011-
Figure 5—source code 1
Pepmatch code Code (developed in C language) for Figure 5.
- https://doi.org/10.7554/eLife.45644.012

Alignment of Hic74 sequences recovered from the Unionoida reference shells and the ornaments.
The reference Hic74 [Hyriopsis cumingii] is shown at the top of the alignment and is highlighted in yellow. Dashes indicate where the sequence was not covered in the samples analysed in this study; amino acid residues highlighted in colour show all disagreements with the reference Hic74 [Hyriopsis cumingii].
-
Figure 6—source data 1
Product ion spectra supporting the amino acid mutations shown in Figure 6.
- https://doi.org/10.7554/eLife.45644.024
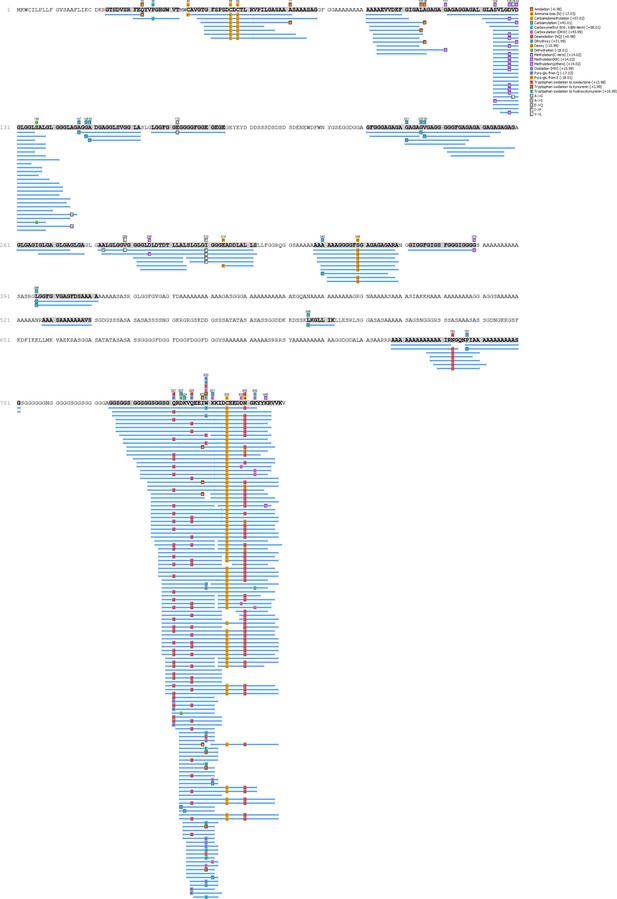
Unio crassus - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.014
Unio pictorum - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.015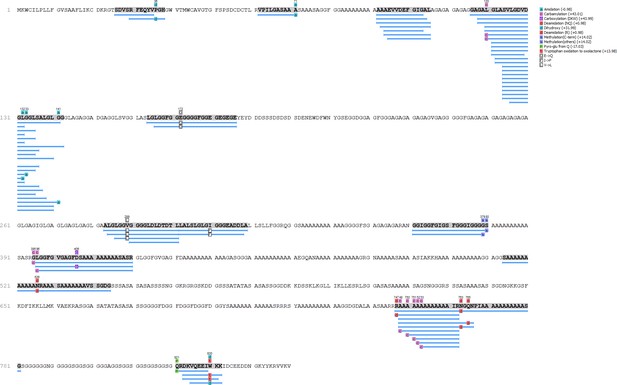
Margaritifera margaritifera - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.016
Pseudunio auricularius - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.017
PesB - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.018
HavA - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.019
HavB - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.020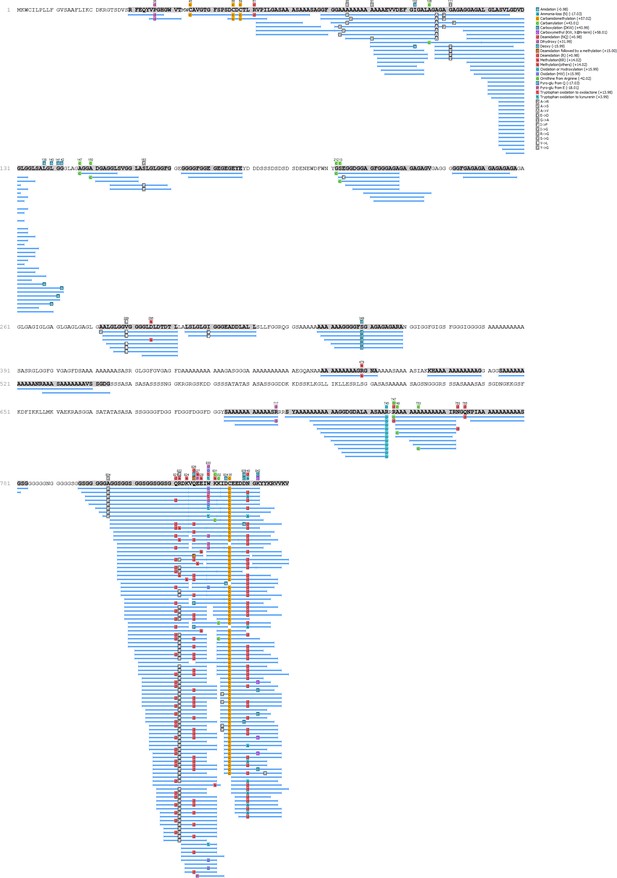
HavC - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.021
HorB - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.022
HorC - Hic74 coverage.
https://doi.org/10.7554/eLife.45644.023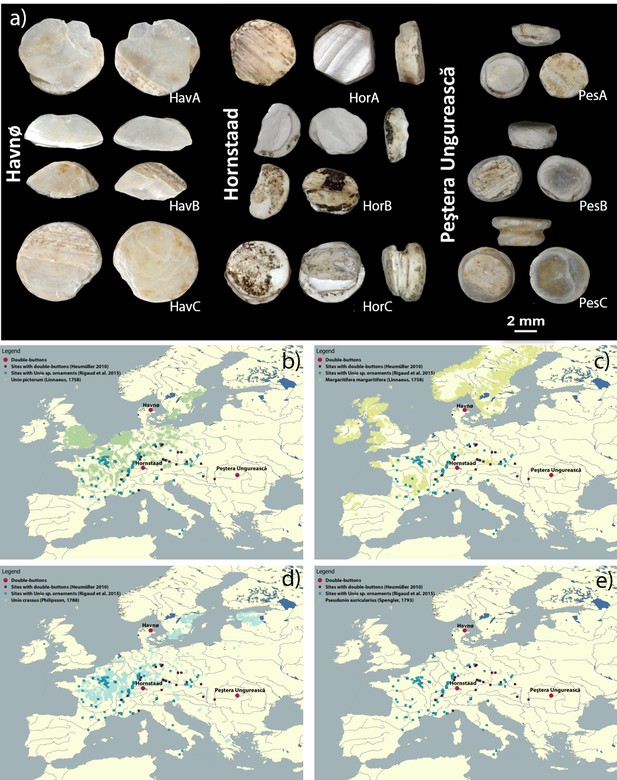
(a) Double-button samples from the archaeological sites of Havnø (Denmark), Hornstaad-Hörnle IA (Germany) and Peştera Ungurească (Romania). Findings of double-buttons and of Unio ornaments as reported in the literature, compared to the present occurrence of Unio pictorum (b), Margaritifera margaritifera (c), Unio crassus (d), Pseudunio auricularius (e) (data obtained from GBIF, the Global Biodiversity Information Facility, GBIF.org, 2018). The three sets of double-buttons (Doppelknöpfe) analysed here come from the archaeological sites of Havnø (Denmark), Hornstaad-Hörnle IA (Germany) and Peştera Ungurească (Romania) and approximately span the period between 4200 and 3800 BCE.

The double-buttons from Havnø.
https://doi.org/10.7554/eLife.45644.029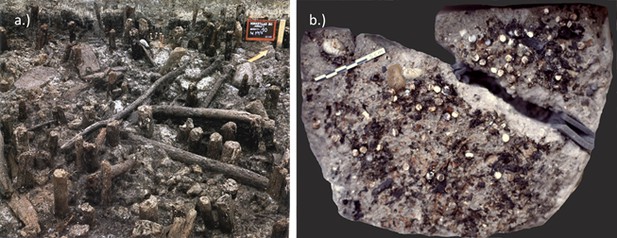
Excavations at Hornstaad D-Hörnle site (a) and the discovery of the double-buttons (b).
https://doi.org/10.7554/eLife.45644.030
Marine and freshwater shells included in this study for comparative analysis: possible sources of raw material used for the manufacture of the double-buttons.
https://doi.org/10.7554/eLife.45644.031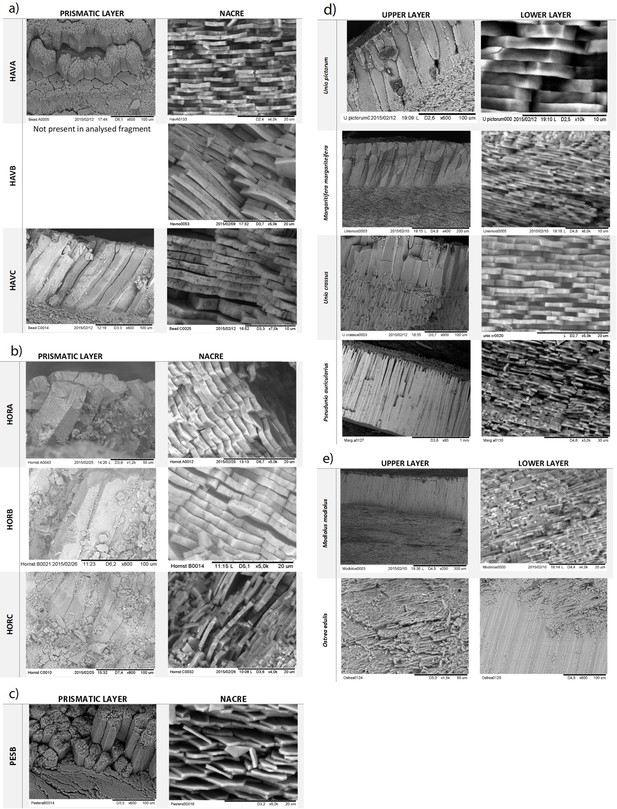
SEM microstructural analysis of archaeological double buttons (a–c) and mollusc shells (d,e).
Double-buttons: a) Havnø (HavA, HavB, HavC), b) Hornstaad (HorA, HorB, HorC), c) Peştera Ungurească (PesB). Reference shells: d) freshwater unionoid shells (modern Unio pictorum, Margaritifera margaritifera and sub-fossil Unio crassus, Pseudunio auricularius), e) marine shells (Modiolus modiolus and Ostrea edulis).
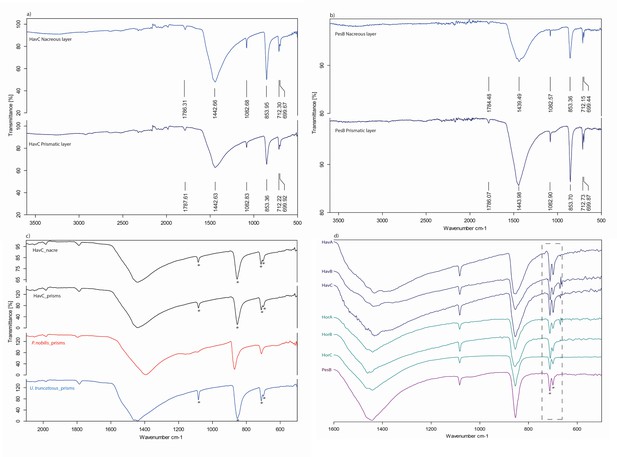
FTIR-ATR spectra of the double-buttons.
Asterisks mark aragonite marker absorption bands. Nacreous and prismatic layers of (a) HavC, (b) PesB; (c) FTIR-ATR spectra comparison between HavC nacre and prismatic layers (black) with calcitic prismatic layer of Pinna nobilis (red) and aragonitic Unio truncatosus (blue), confirming the fully aragonitic mineralogy of both layers; (d) FTIR-ATR spectra of all the double-buttons, sampled in ‘bulk’: the presence of doublets (CO in-plane bending mode) at ~ 712 and~700 cm−1 (dashed line) in all samples indicates absence of recrystallization of the biogenic carbonate.
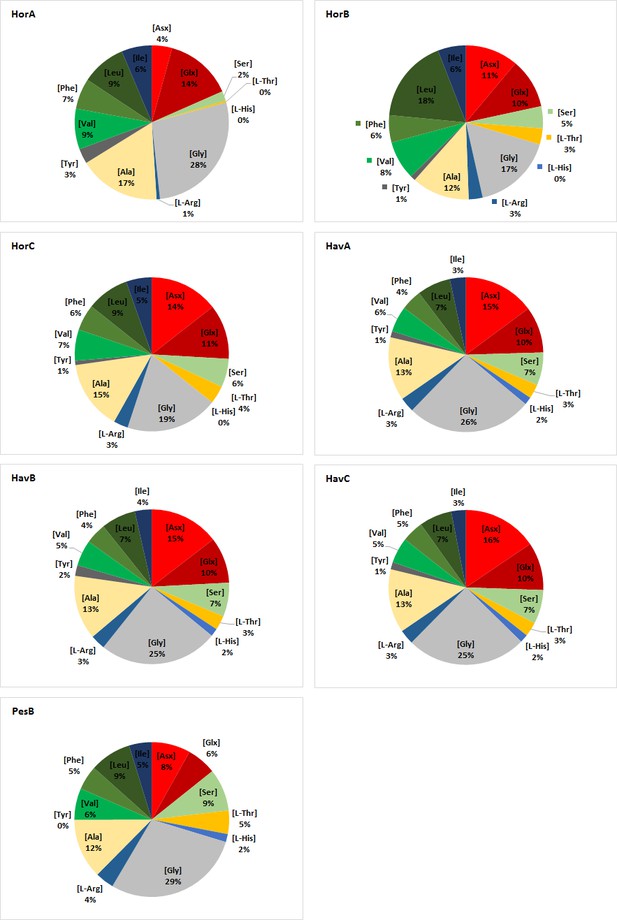
Relative THAA composition of double-button samples.
https://doi.org/10.7554/eLife.45644.037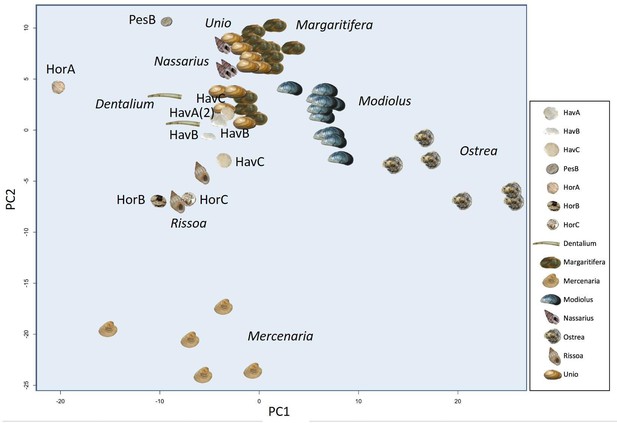
Principal Component Analysis (PCA) plot showing the similarity or differences between the amino acid composition of double-buttons and a range of shell taxa (reference taxa from Demarchi et al., 2014).
https://doi.org/10.7554/eLife.45644.038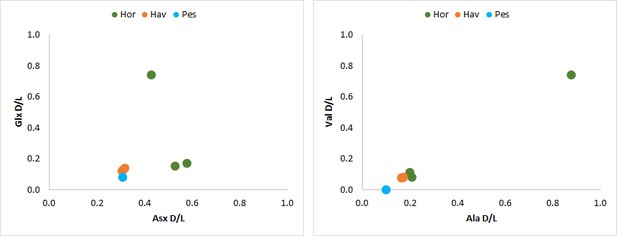
Total hydrolysable amino acid D/L values for all double-buttons (Glx vs Asx, left; Val vs Ala, right).
https://doi.org/10.7554/eLife.45644.039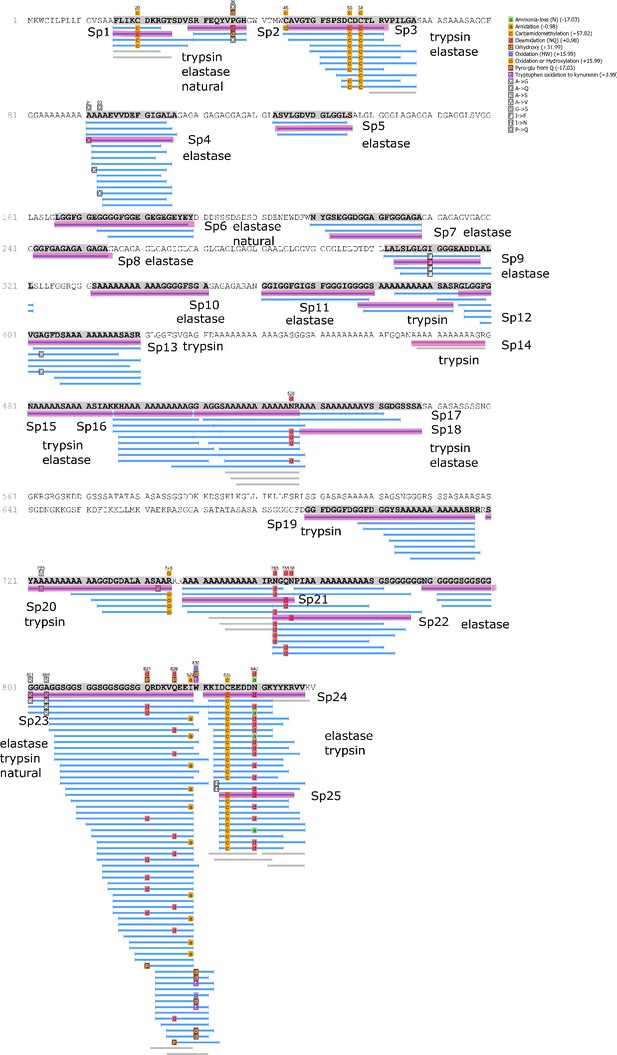
Protein Hic74 identified in Unio pictorum: sequence coverage, highlighting in pink the product ion spectra ('Sp') shown below.
Sequences reconstructed by assisted de novo on the basis of mono-charged ions mainly (spectra were acquired on the 400-1600 m/z range and multiply-charged ions were detected).
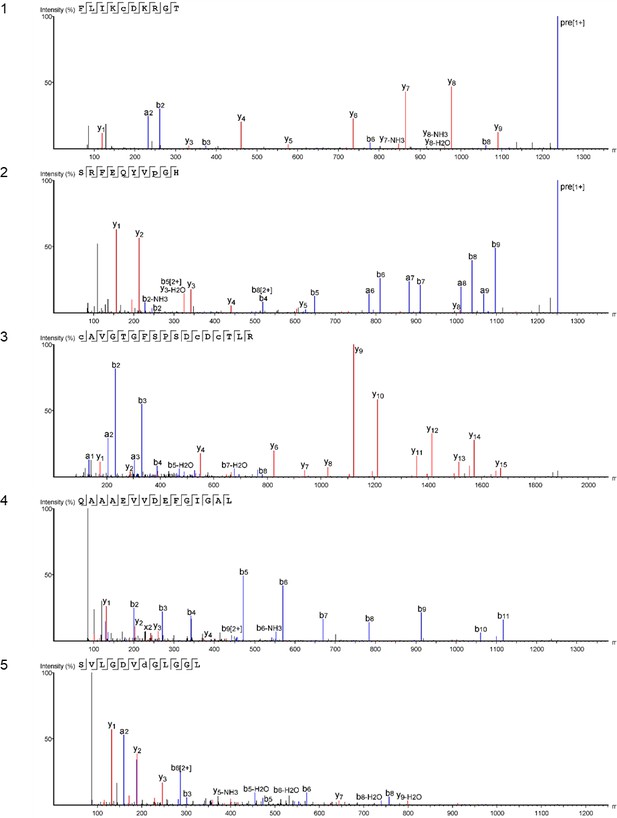
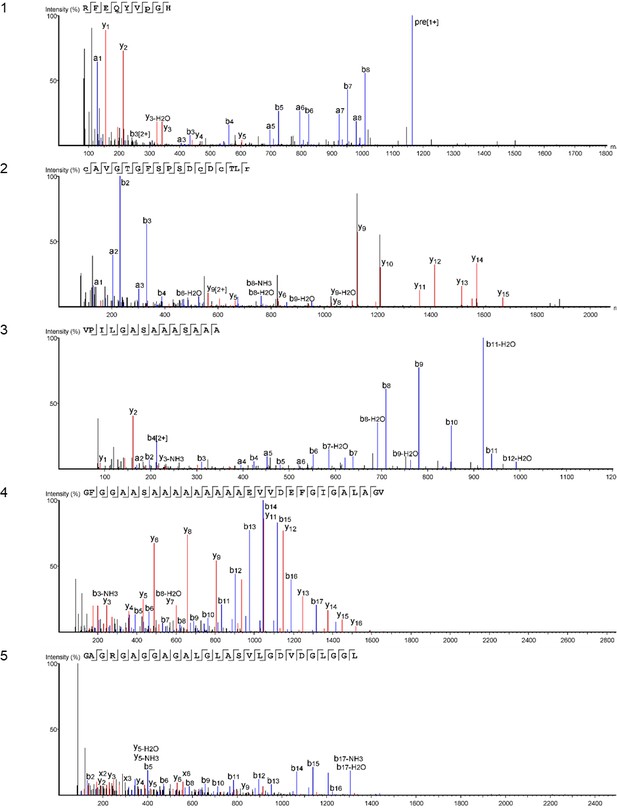
Product ion spectra [1 - 5] supporting the coverage of protein Hic74 identified in Unio pictorum.
https://doi.org/10.7554/eLife.45644.042
Product ion spectra [6 - 10] supporting the coverage of protein Hic74 identified in Unio pictorum.
https://doi.org/10.7554/eLife.45644.043
Product ion spectra [11 - 15] supporting the coverage of protein Hic74 identified in Unio pictorum.
Product ion spectra [16 - 20] supporting the coverage of protein Hic74 identified in Unio pictorum.
https://doi.org/10.7554/eLife.45644.045
Product ion spectra [21 - 25] supporting the coverage of protein Hic74 identified in Unio pictorum.
https://doi.org/10.7554/eLife.45644.046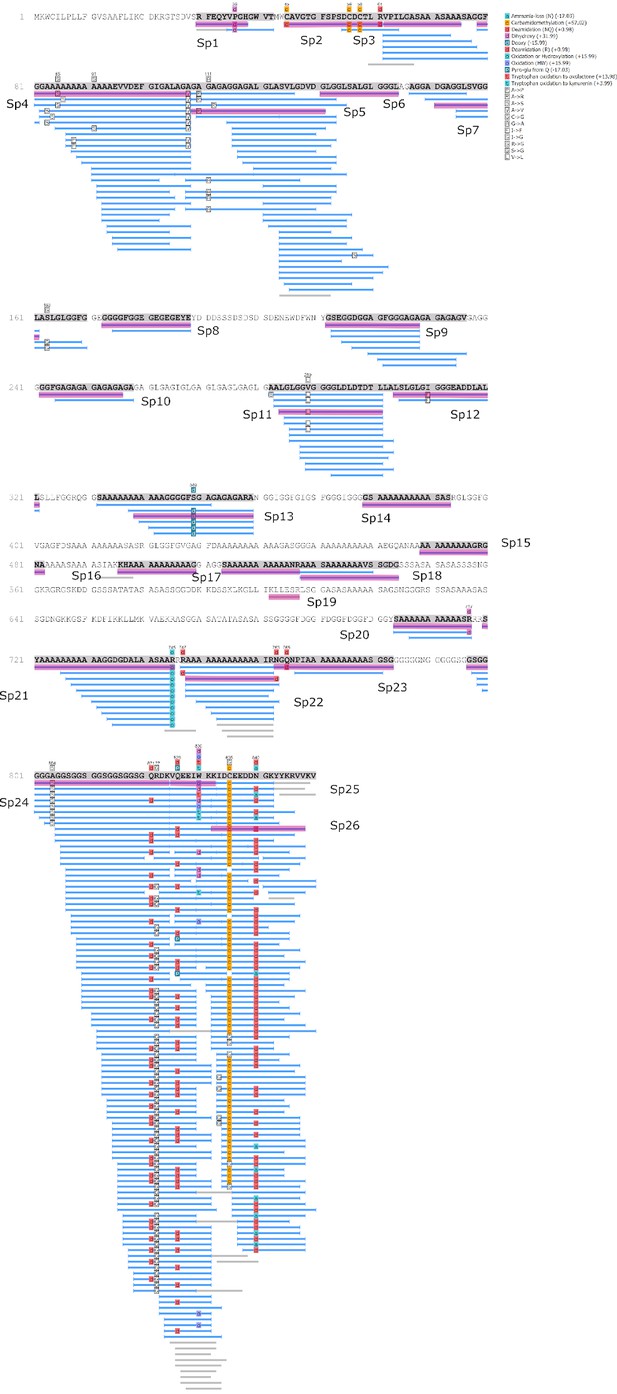
Protein Hic74 identified in the double-button HavC: sequence coverage, highlighting in pink the product ion spectra ('Sp') shown below.
Sequences reconstructed by assisted de novo on the basis of mono-charged ions mainly (spectra were acquired on the 400-1600 m/z range and multiply-charged ions were detected).
Product ion spectra [1 - 5] supporting the coverage of protein Hic74 identified in the double-button HavC.
https://doi.org/10.7554/eLife.45644.049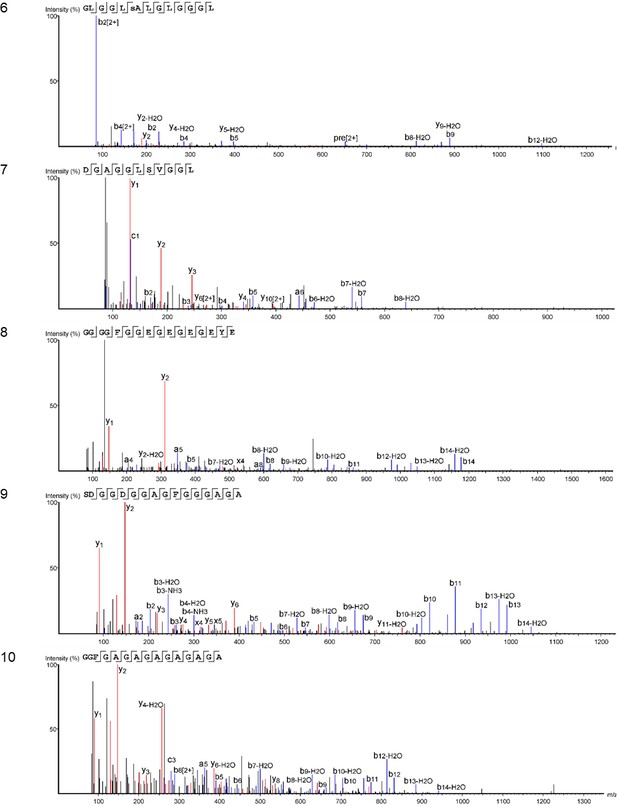
Product on spectra [6 -10] supporting the coverage of protein Hic74 identified in the double-button HavC.
https://doi.org/10.7554/eLife.45644.050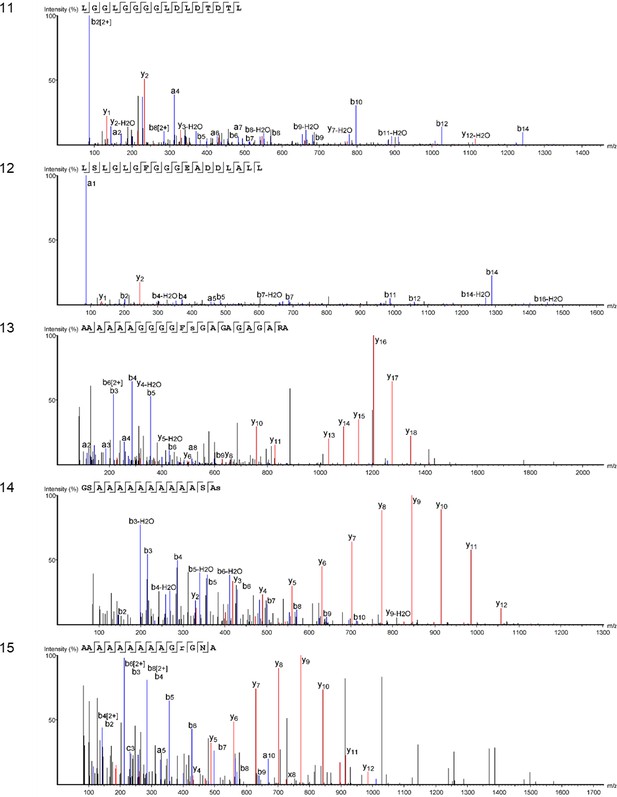
Product ion spectra [11 - 15] supporting the coverage of protein Hic74 identified in the double-button HavC.
https://doi.org/10.7554/eLife.45644.051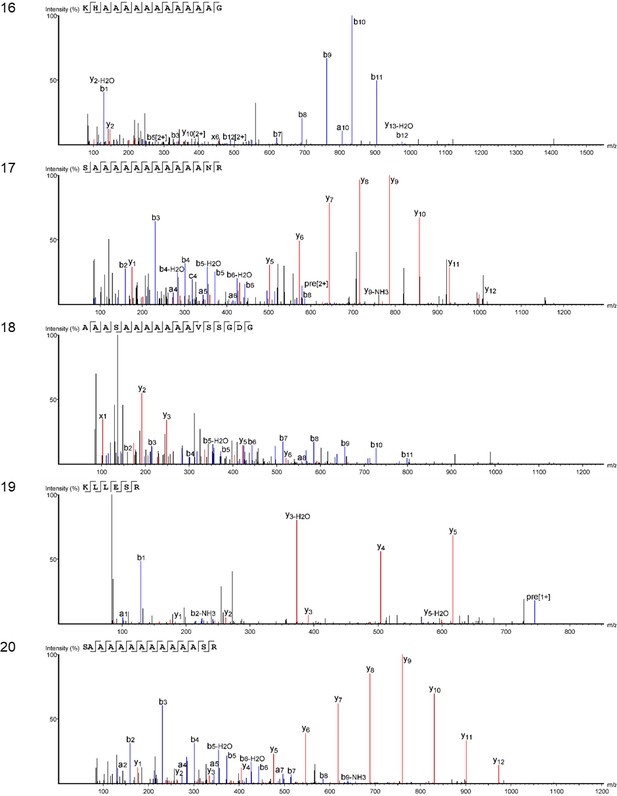
Product ion spectra [16 - 20] supporting the coverage of protein Hic74 identified in the double-button HavC.
https://doi.org/10.7554/eLife.45644.052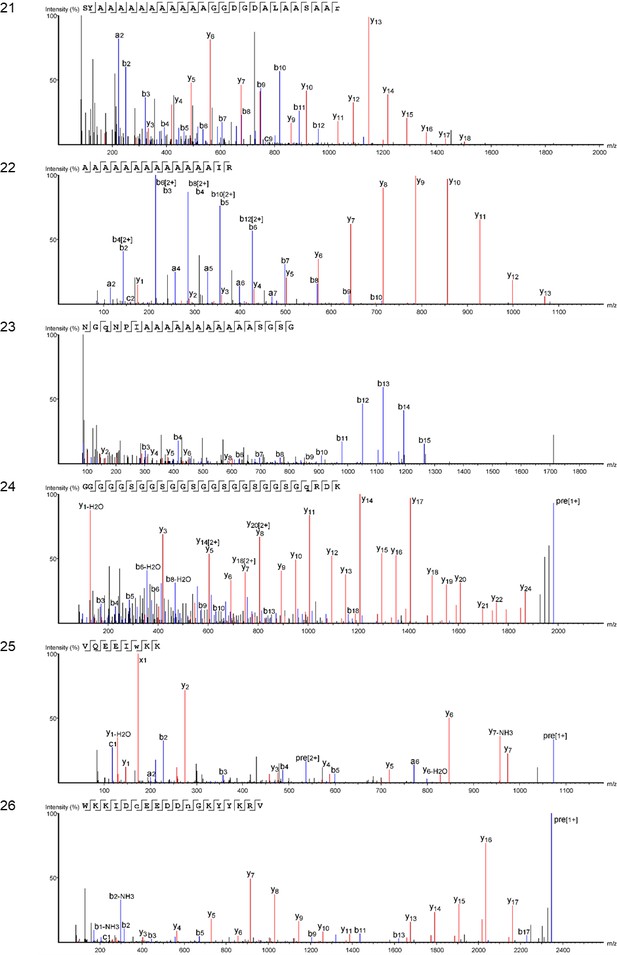
Product ion spectra [21 - 26] supporting the coverage of protein Hic74 identified in the double-button HavC.
https://doi.org/10.7554/eLife.45644.053
Marine shell proteins identified in double-button HavB: (a) MSI60-related protein (Pinctada fucata); (b) Precollagen D (Mytilus edulis).
Note that both are supported only by repetitive low complexity (RLC) domains. Sequences reconstructed by assisted de novo on the basis of mono-charged ions mainly (spectra were acquired on the 400–1600 m/z range and multiply-charged ions were detected).
Circular diagram representing the extent of similarity between the proteomes of six reference mollusc shells based on the identified EST sequences.
https://doi.org/10.7554/eLife.45644.055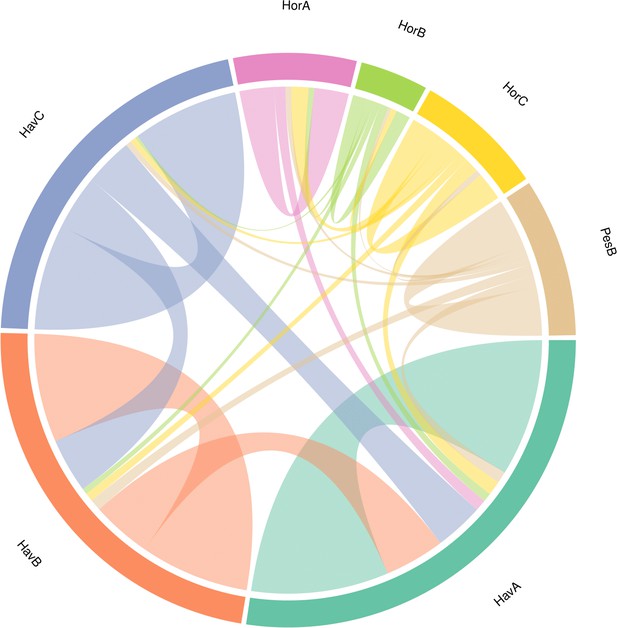
Circular diagram representing the similarity between the proteomes of the seven double-buttons based on the identified EST sequences.
https://doi.org/10.7554/eLife.45644.056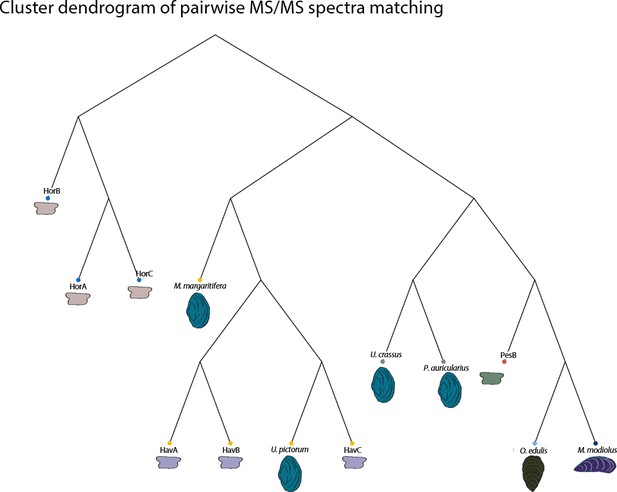
Pairwise MS/MS comparison of the seven archaeological double-buttons and six reference shells (freshwater and marine): the cluster dendrogram is obtained from a distance matrix from proteome-wide distance calculations of product ion spectra implemented in R using the DISMS2 code.
https://doi.org/10.7554/eLife.45644.057Tables
Summary of the materials analysed in this study (archaeological ornaments and reference shells) and information on their context, chronology and taxonomic determination.
https://doi.org/10.7554/eLife.45644.005| Sample type | Site | Cultural group | Time span | Taxonomic determination before this study | Other molluscan fauna present at the site |
|---|---|---|---|---|---|
| Double-buttons | Havnø | Ertebølle | 4200–4000 cal BCE (radiocarbon dating of the Late Mesolithic horizon; Andersen, 2000; Andersen, 2008) | Unknown, presumed marine shell | Abundant edible marine shells: Ostrea edulis, Littorina sp., Mytilus edulis, Cerastoderma edule (Andersen, 2000; Andersen, 2008) |
| Hornstaad-Hörnle IA | Hornstaad group (early phase of the regional Late Neolithic) | 3917–3902 BCE (dendrochronology; Billamboz, 2006) | Debated: marine (Ostrea edulis) vs freshwater (Margaritifera margaritifera; Heumüller, 2010; Borrello and Girod, 2008) | Mediterranean marine shells (exotic, non edible): Columbella rustica (Borrello and Girod, 2008), Callista chione, Astarte borealis, Dentalium vulgare (Heumüller, 2010) | |
| Peştera Ungurească | Toarte Pastilate and transition to Coţofeni | 4260–3820 cal BCE (range of radiocarbon dates, at 1σ, of layers 2B, 2A3 and 2A, Toarte Pastilate)(Biagi and Voytek, 2006) | Unio cf. crassus (Girod, 2010a, Figure 6B) | Abundant terrestrial taxa (naturally occurring). Occasional presence of freshwater species (Anisus spirorbis, Pisidium milium, Lithoglyphus naticoides, Lymnaea truncatula, Planorbis cf. carinatus; Girod, 2010a) | |
| Reference shell | Limfjord (Northern Jutland) | Modern | Ostrea edulis | ||
| Limfjord (Northern Jutland) | Modern | Modiolus modiolus | |||
| Limfjord (Northern Jutland) | Modern | Margaritifera margaritifera (determined by F.M.) | |||
| France (Izeure) | Modern | Unio pictorum | |||
| Peştera Ungurească | Toarte Pastilate and transition to Coţofeni | 4260–3820 cal BCE (range of radiocarbon dates, at 1σ, of layers 2B, 2A3 and 2A, Toarte Pastilate) (Biagi and Voytek, 2006) | Unio cf. crassus (Girod, 2010a) | Abundant terrestrial taxa (environmental signal). Occasional occurrence of freshwater species (Anisus spirorbis, Pisidium milium, Lithoglyphus naticoides, Lymnaea truncatula, Planorbis cf. carinatus). Not suitable as raw material for the double-buttons (Girod, 2010a) | |
| Isorella | Vhò | 5226–5023 cal BCE at 2σ (Starnini et al., 2018) | Pseudunio auricularius (Biddittu and Girod, 2003; Girod, 2010b) | Marine taxa (typically used as ornaments): C. rustica, Spondylus (fragment of a bracelet; Girod, 2010b) |
Main protein sequences identified in the double-buttons from Havnø, Hornstaad-Hörnle IA and Peştera Ungurească and their presence/absence in the analysed set of reference freshwater and marine shells (black dots).
Numbers indicate total number of peptide sequences identified and the cell colour is proportional to the coverage of the sequence itself. Threshold values for peptide and protein identification: false discovery rate (protein FDR) = 0.5%, protein score −10lgP ≥ 40, unique peptides ≥ 2, de novo sequences scores (ALC%) ≥ 50. Asterisks (*) indicate proteins identified only when using less stringent parametres: protein score −10lgP ≥ 20; unique peptides ≥ 1. Note that molecular sequence databases for molluscan species are incomplete and biased towards well-studied model organisms. The peptide sequences recovered in our study were identified using sequence homologies with proteins originally described from Hyriopsis cumingii, Crassostrea sp., Pinctada sp., Mytilus sp. and several others. As a result of database insufficiency, the bioinformatic search of these ‘shellomes’ could not identify the exact taxon of our samples, but provided a strong indication of the fact that the closest taxon to that of the ornaments (and of the freshwater reference shells) is the pearl-producing triangle sail mussel Hyriopsis cumingii (Unionoida).
| Proteins present in database from | Identified proteins | Double-buttons | Freshwater | Marine | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Order | Genus | HavA | HavB | HavC | HorA | HorB | HorC | PesB | U.p | U.c | M.m | P.a | Mo.M | O.e | |
| Unionoida | Hyriopsis | Hic74 [Hyriopsis cumingii] | 132 | 158 | 260 | 6 | 11 | 21 | ● | ● | ● | ● | |||
| Hic52 nacreous layer matrix protein [Hyriopsis cumingii] | 1* | 1* | 2* | ● | ● | ● | ● | ||||||||
| Silkmapin (isoforms: nasilin 1 and nasilin 2) [Hyriopsis cumingii] | 1* | 3 | 3 | 5 | ● | ● | ●* | ●* | |||||||
| Ostreida | Pinctada | MSI60-related protein [Pinctada fucata] | 6 | 27 | 12 | ● | ● | ● | ● | ||||||
| Insoluble matrix protein [Pinctada fucata] | 4 | 33 | ● | ● | ● | ||||||||||
| Crassostrea | Glycine-rich cell wall structural protein-like [Crassostrea virginica/gigas] | 17 | 11 | 12 | 14 | ● | |||||||||
| Glycine-rich protein 23-like [Crassostrea virginica] | 8 | 11 | 6 | ● | ● | ||||||||||
| Antifreeze protein Maxi-like [Crassostrea virginica] | 4 | 4 | ● | ||||||||||||
| Mytilida | Bathymodiolus | MSI60-related protein partial [Bathymodiolus platifrons] | 6 | 11 | ● | ● | ● | ||||||||
| Mytilus | Precollagen D [Mytilus edulis] | 16 | 26 | 23 | 9 | ● | ● | ● | |||||||
| Nongradient byssal precursor [Mytilus edulis] | 10 | 10 | 25 | ● | |||||||||||
| Other | Other | Predicted: transcription factor hamlet-like partial [Octopus bimaculoides] | 5 | 6 | 11 | ● | ● | ||||||||
| Hypothetical protein OCBIM_22008720 mg partial [Octopus bimaculoides] | 6 | 11 | |||||||||||||
| Coverage | ≥55% | ≥35% | ≥15% | ≥10% | ≥1% | Presence ● | |||||||||
-
Table 2—source data 1
Palaeoshellomics.
The complete proteomics dataset obtained on reference shells and archaeological ornaments
- https://doi.org/10.7554/eLife.45644.008
Potential amino acid substitutions detected in the samples analysed in this study, compared to the reference Hic74 sequence [Hyriopsis cumingii].
Positions are derived from the sequence alignment shown in Figure 6. Dashes indicate that the position was not covered for that sample; question mark symbols indicate ambiguous substitutions. Hic74 coverages for each sample and supporting product ion spectra are presented in Figure 6—figure supplement 1, Figure 6—figure supplement 2, Figure 6—figure supplement 3, Figure 6—figure supplement 4, Figure 6—figure supplement 5, Figure 6—figure supplement 6, Figure 6—figure supplement 7, Figure 6—figure supplement 8, Figure 6—figure supplement 9, Figure 6—figure supplement 10, as well as in Figure 6—source data 1.
| Hic74 [Hyriopsis cumingii] | A | A | A | A | A | A | A | G | D | G | S | E | G | A | A | L | V | G | L | I | A | G | A | Q | R | E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA position | 83 | 85 | 91 | 93 | 106 | 108 | 110 | 111 | 151 | 152 | 163 | 172 | 175 | 282 | 283 | 284 | 289 | 292 | 306 | 310 | 403 | 801 | 804 | 821 | 822 | 827 |
| U. crassus | - | - | A | A | A | A | A | G | D | G | - | Q | G | A | ? | L | ? | G | L | F | A | - | - | Q | R | E |
| U. pictorum | - | - | ? | ? | ? | - | - | - | - | - | - | Q | G | - | - | - | - | - | L | F | ? | S | S | Q | R | E |
| M. margaritifera | - | - | - | A | - | - | - | - | - | - | - | Q | G | - | A | L | L | G | L | F | A | - | - | Q | R | E |
| P. auricularius | - | A | A | A | A | A | A | - | ? | ? | D | E | L | - | A | F | V | E | F | I | - | - | - | H | H | D |
| HavA | A | S | A | A | A | ? | ? | A | - | - | - | Q | G | A | A | L | L | G | L | F | - | - | - | Q | G | E |
| HavB | ? | S | A | A | A | V | - | - | - | - | - | - | - | - | A | L | L | G | L | F | - | - | - | Q | G | E |
| HavC | ? | S | A | A | A | V | ? | A | D | G | G | - | G | ? | A | L | L | G | L | F | - | G | S | Q | G | E |
Stable isotope composition of the biogenic carbonate of the double-buttons.
https://doi.org/10.7554/eLife.45644.034| Sample | δ13C (‰) | δ18O (‰) |
|---|---|---|
| HavA | −11.8 ± 0.19 | −5.0 ± 0.09 |
| HavB | −10.9 ± 0.09 | −5.7 ± 0.17 |
| HavC | −10.6 ± 0.07 | −5.3 ± 0.08 |
| HorA | −11.7 ± 0.07 | −9.8 ± 0.11 |
| HorB | −11.3 ± 0.07 | −9.2 ± 0.07 |
| HorC | −8.7 ± 0.07 | −8.9 ± 0.06 |
| PesB_n | −13.1 ± 0.01 | −6.8 ± 0.03 |
| PesB_p | −10.7 ± 0.01 | −5.5 ± 0.03 |
Total hydrolysable amino acid (THAA) concentrations measured in archaeological double-button samples (pmol/mg).
Average and standard deviation were calculated on two analytical replicates. Values for Havnø include the average and standard deviation for the two subsamples taken from each double-button.
| [Asx] | [Glx] | [Ser] | [Gly] | [Ala] | [Val] | [Phe] | [Ile] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AV | SD | AV | SD | AV | SD | AV | SD | AV | SD | AV | SD | AV | SD | AV | SD | |
| HOR-A | 150 | 1 | 497 | 6 | 72 | 0 | 965 | 36 | 604 | 0 | 300 | 1 | 227 | 2 | 223 | 3 |
| HOR-B | 340 | 128 | 322 | 126 | 142 | 47 | 516 | 301 | 372 | 167 | 250 | 40 | 175 | 39 | 181 | 32 |
| HOR-C | 429 | 61 | 340 | 11 | 177 | 25 | 574 | 68 | 434 | 46 | 194 | 43 | 163 | 11 | 160 | 19 |
| HAV-A | 723 | 48 | 470 | 17 | 334 | 12 | 1274 | 82 | 644 | 57 | 262 | 25 | 212 | 20 | 163 | 11 |
| HAV-B | 711 | 87 | 470 | 45 | 346 | 25 | 1214 | 176 | 656 | 66 | 264 | 24 | 213 | 18 | 173 | 18 |
| HAV-C | 703 | 14 | 452 | 13 | 321 | 21 | 1132 | 228 | 602 | 19 | 241 | 5 | 204 | 6 | 138 | 8 |
| PES-B | 452 | 8 | 344 | 13 | 494 | 1 | 1605 | 66 | 699 | 1 | 368 | 1 | 282 | 1 | 264 | 13 |
Total hydrolyzable amino acid (THAA) D/L values measured in archaeological double-buttons.
Average and standard deviation were calculated on two analytical replicates. Values for Havnø include the average and standard deviation for the two subsamples taken from each double-button.
| Asx D/L | Glx D/L | Ser D/L | Ala D/L | |||||
|---|---|---|---|---|---|---|---|---|
| AV | SD | AV | SD | AV | SD | AV | SD | |
| HOR-A | 0.430 | 0.000 | 0.740 | 0.003 | 0.000 | 0.000 | 0.880 | 0.001 |
| HOR-B | 0.530 | 0.073 | 0.150 | 0.421 | 0.720 | 0.515 | 0.200 | 0.477 |
| HOR-C | 0.580 | 0.033 | 0.170 | 0.016 | 0.760 | 0.026 | 0.210 | 0.010 |
| HAV-A | 0.313 | 0.015 | 0.128 | 0.032 | 0.455 | 0.030 | 0.165 | 0.013 |
| HAV-B | 0.318 | 0.010 | 0.135 | 0.010 | 0.480 | 0.024 | 0.168 | 0.010 |
| HAV-C | 0.308 | 0.010 | 0.120 | 0.008 | 0.475 | 0.026 | 0.175 | 0.006 |
| PES-B | 0.310 | 0.000 | 0.080 | 0.000 | 0.280 | 0.000 | 0.100 | 0.000 |
| Val D/L | Phe D/L | Ile D/L | ||||||
| AV | SD | AV | SD | AV | SD | |||
| HOR-A | 0.740 | 0.001 | 0.730 | 0.004 | 0.850 | 0.029 | ||
| HOR-B | 0.110 | 0.449 | 0.290 | 0.308 | 0.140 | 0.488 | ||
| HOR-C | 0.080 | 0.019 | 0.320 | 0.026 | 0.100 | 0.039 | ||
| HAV-A | 0.073 | 0.015 | 0.180 | 0.008 | 0.045 | 0.052 | ||
| HAV-B | 0.075 | 0.013 | 0.193 | 0.013 | 0.065 | 0.044 | ||
| HAV-C | 0.078 | 0.010 | 0.170 | 0.008 | 0.023 | 0.045 | ||
| PES-B | 0.000 | 0.000 | 0.140 | 0.000 | 0.000 | 0.000 | ||
Mass of biogenic carbonates analysed for proteomics
https://doi.org/10.7554/eLife.45644.040| Sample | Powder mass (mg) | |
|---|---|---|
| Archaeological samples | HavA | 92.9 |
| HavB | 69.6 | |
| HavC | 172.5 | |
| HorA | 49.23 | |
| HorB | 47.34 | |
| Horc | 47.23 | |
| PesB | 32.48 | |
| Sample | Mass (g) | |
| Freshwater unionoid shells | U. pictorum | 10 |
| U. crassus | 3 | |
| M. margaritifera | 10 | |
| P. auricularius | 3 | |
| Marine shells | O. edulis | 10 |
| M. modiolus | 10 |
Extent of Asn (N) and Gln (Q) deamidation (N→D; Q→E) in the peptides identified in the Hic74 sequence.
https://doi.org/10.7554/eLife.45644.047| Hav A | Hav B | Hav C | Hor A | Hor B | Hor C | Pes B | U. pict orum | U. cras sus | M. marga ritifera | P. auricu larius | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| # Q | 28 | 105 | 196 | - | - | - | 12 | 136 | 237 | 13 | 37 |
| # N | 38 | 31 | 104 | 4 | 71 | 96 | 8 | 88 | |||
| # Q→E | 3 | 33 | 71 | 1 | 18 | 67 | 1 | 12 | |||
| # N→D | 19 | 17 | 53 | 2 | 28 | 66 | 3 | 57 | |||
| % Q→E | 11 | 36 | 36 | 8 | 13 | 28 | 8 | 32 | |||
| % N→D | 50 | 51 | 51 | 50 | 39 | 69 | 37 | 65 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45644.026





