Gene activation by a CRISPR-assisted trans enhancer
Figures
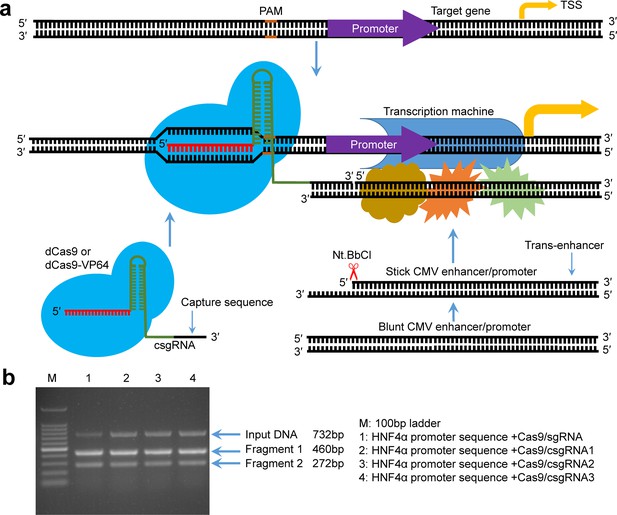
Principle of gene expression activation by the CRISPR-assisted trans enhancer and evaluation of designed csgRNAs.
(a) Schematic illustration of the principle of gene expression activation by the CRISPR-assisted trans enhancer. A capture sequence is added to the 3′ end of sgRNA, which is used to capture a trans CMV enhancer with a single-stranded overhang that can anneal with the capture sequence of sgRNA. The captured trans CMV enhancer may function like the natural looped cis enhancer to activate transcription of the gene of interest, including exogenous and endogenous genes. (b) In vitro target DNA cutting by the Cas9-csgRNA complex. DNA fragments (732 bp) amplified from the HNF4α promoter region were, respectively, cut by the Cas9/csgRNA and Cas9/sgRNA complexes. csgRNA1, csgRNA2 and csgRNA3 had the same target sequence but different capture sequences.
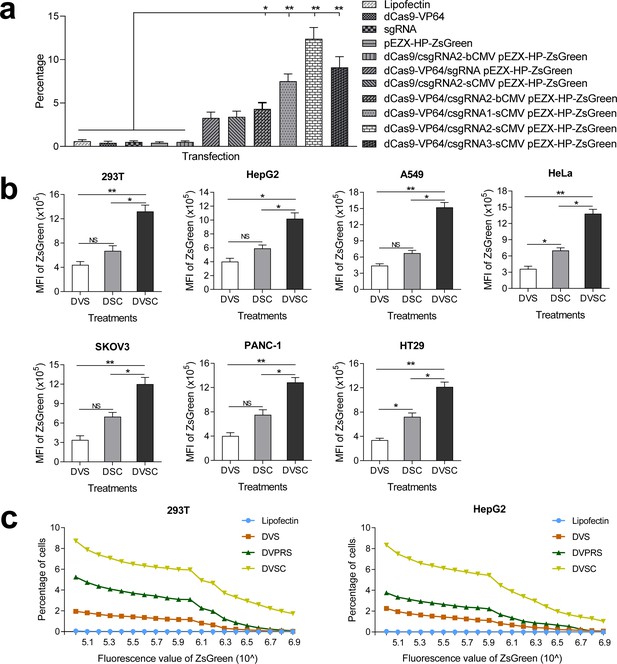
Activation of an exogenous reporter gene ZsGreen under the control of a HNF4α promoter by the CRISPR-assisted trans enhancer in multiple cells.
(a) Transcriptional activation of reporter gene ZsGreen in various cells transfected by different vectors. The florescence intensity of cells was analyzed by flow cytometry and is shown as the mean fluorescence intensity (MFI). Transfections: DVS, dCas9-VP64/sgRNA; DSC, dCas9/csgRNA-sCMV; DVSC, dCas9-VP64/csgRNA-sCMV. (b) Comparison between trans enhancer and VPR. Cells were transfected with three different transcriptional activation systems to activate reporter gene ZsGreen. The florescence intensity of cells was analyzed by flow cytometry and the number of cells with certain fluorescence intensity was counted. Transfections: Lipo, lipofectin; DVS, dCas9-VP64/sgRNA; DVPRS; dCas9-VPR/csgRNA; DVSC, dCas9-VP64/csgRNA-sCMV.
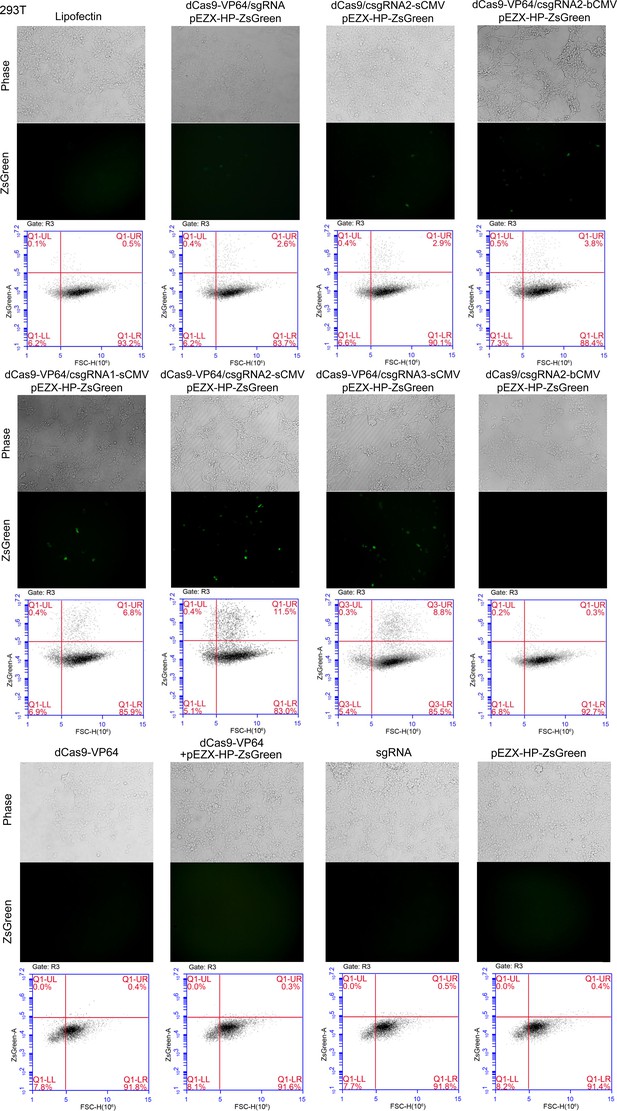
Activation of an exogenous reporter gene ZsGreen under the control of a HNF4α promoter by the CRISPR-assisted trans enhancer in 293 T cells.
Cells were transfected by various vectors. Cells were photographed with a fluorescent microscope and their florescence was analyzed by flow cytometry. The reporter gene activation efficiency was indicated by the percentage of cells with green fluorescence over the threshold (cells in Q1-UR quadrant).
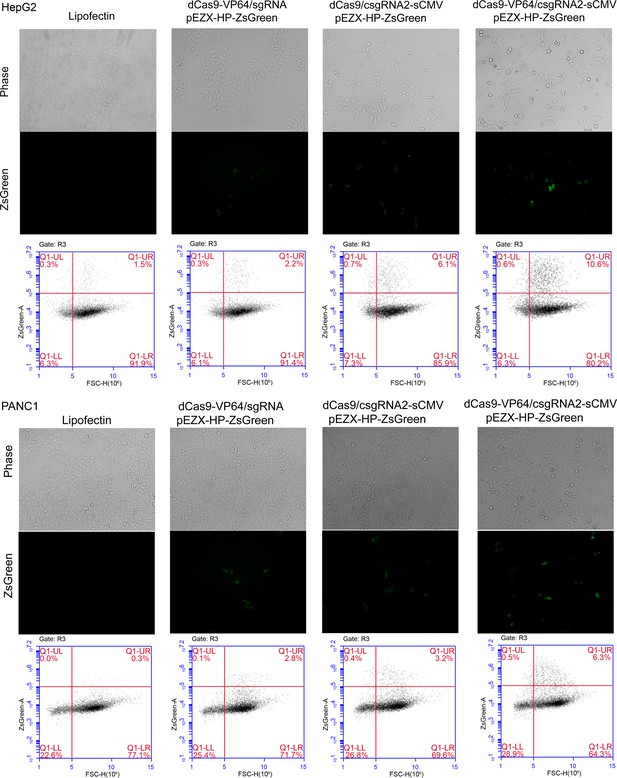
Activation of an exogenous reporter gene ZsGreen under the control of a HNF4α promoter by the CRISPR-assisted trans enhancer in HepG2 and PANC1 cells.
https://doi.org/10.7554/eLife.45973.005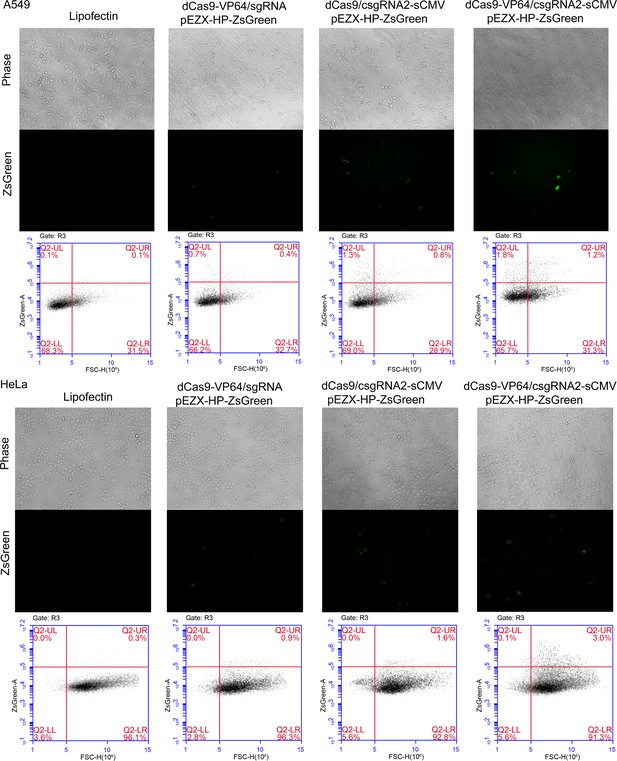
Activation of an exogenous reporter gene ZsGreen under the control of a HNF4α promoter by the CRISPR-assisted trans enhancer in A549 and HeLa cells.
https://doi.org/10.7554/eLife.45973.006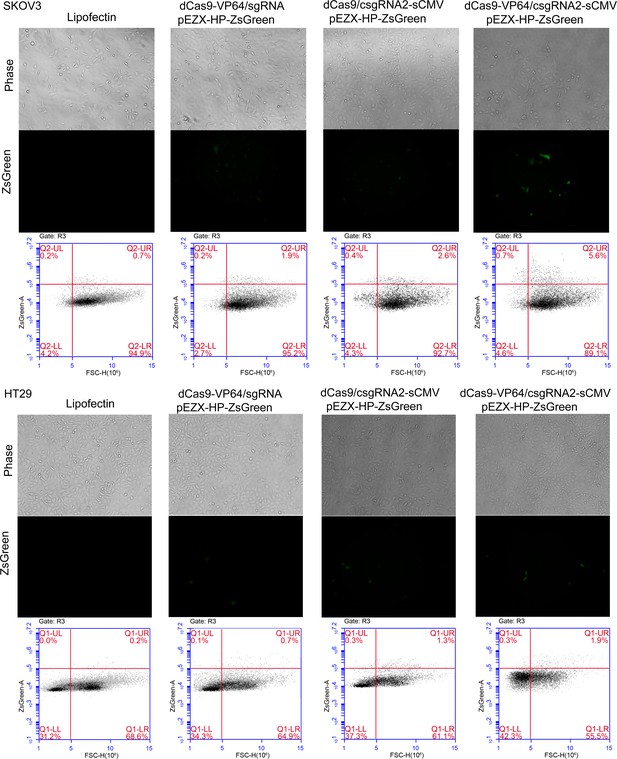
Activation of an exogenous reporter gene ZsGreen under the control of a HNF4α promoter by the CRISPR-assisted trans enhancer in SKOV3 and HT29 cells.
https://doi.org/10.7554/eLife.45973.007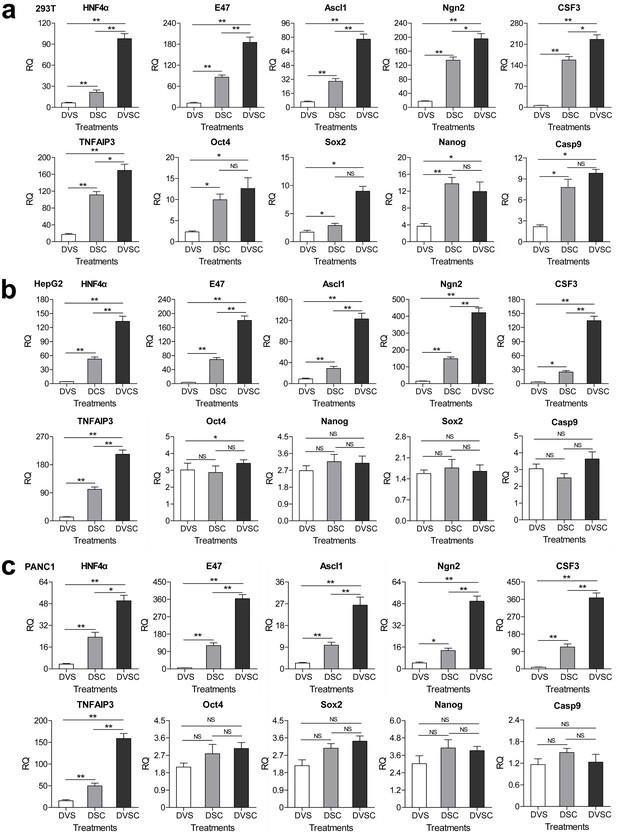
Transcriptional activation of endogenous genes by the CRISPR-assisted trans enhancer.
294T, HepG2 and PANC1 cells were transfected with three different transcriptional activation systems to activate expression of 10 endogenous genes. Gene transcription was detected by qPCR and the expression level is shown as the relative RNA expression fold to house-keeping gene GAPDH. Data are shown as mean ± SD, n = 3. The statistical difference was analyzed using the Student’s t test. *, p<0.05; **, p<0.01; NS, no significant statistical difference. Transfection: DVS, dCas9-VP64/sgRNA; DSC, dCas9/csgRNA2-sCMV; DVSC, dCas9-VP64/csgRNA2-sCMV.
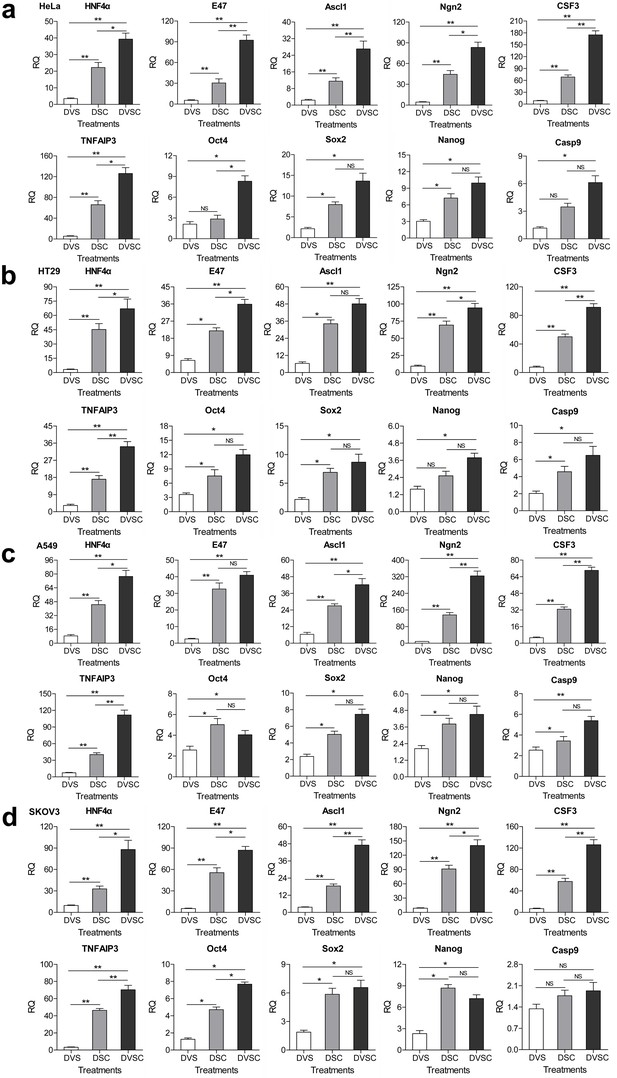
Transcriptional activation of endogenous genes by the CRISPR-assisted trans enhancer.
A549, HeLa, SKOV, and HT29 cells were transfected with three different transcriptional activation systems to activate the expression of 10 endogenous genes.
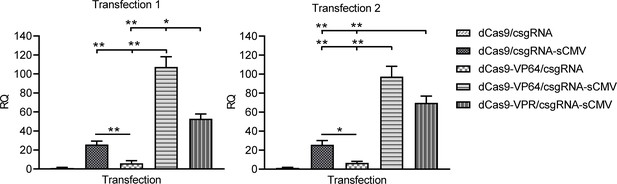
Activation of endogenous HNF4α gene in 293 T cell with trans enhancers based on dCas9-VP64 and dCas9-VPR.
The 293 T cell was transfected with various vectors to activate the endogenous HNF4α gene. The HNF4α and GAPDH genes were detected with qPCR.
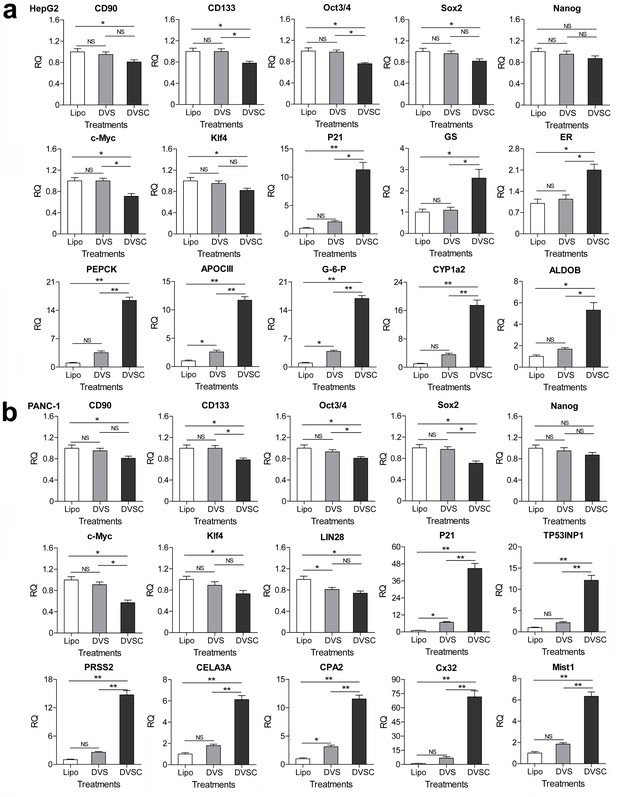
Changes of gene expression in the HNF4α-activated HepG2 cells and E47-activated PANC-1 cells.
(a and b) Changes of gene expression in the HNF4α-activated HepG2 cells (a) and E47-activated PANC-1 cells (b). The gene transcription was detected by qPCR and the expression level is shown as the relative RNA expression fold to house-keeping gene GAPDH. Data are shown as mean ± SD, n = 3. The statistical difference was analyzed by Student’s t test. *, p<0.05; **, p<0.01; NS, no significant statistical difference. Transfection: Lipo, lipofectin; DVS, dCas9-VP64/sgRNA; DVSC, dCas9-VP64/csgRNA2-sCMV.
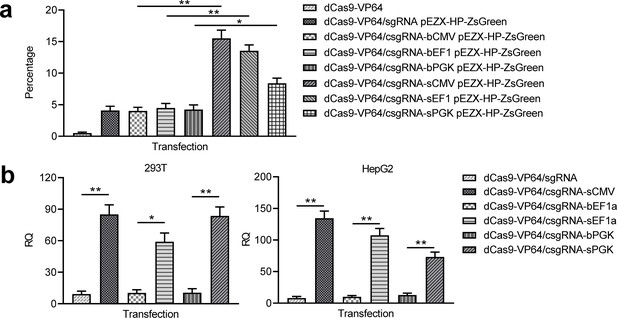
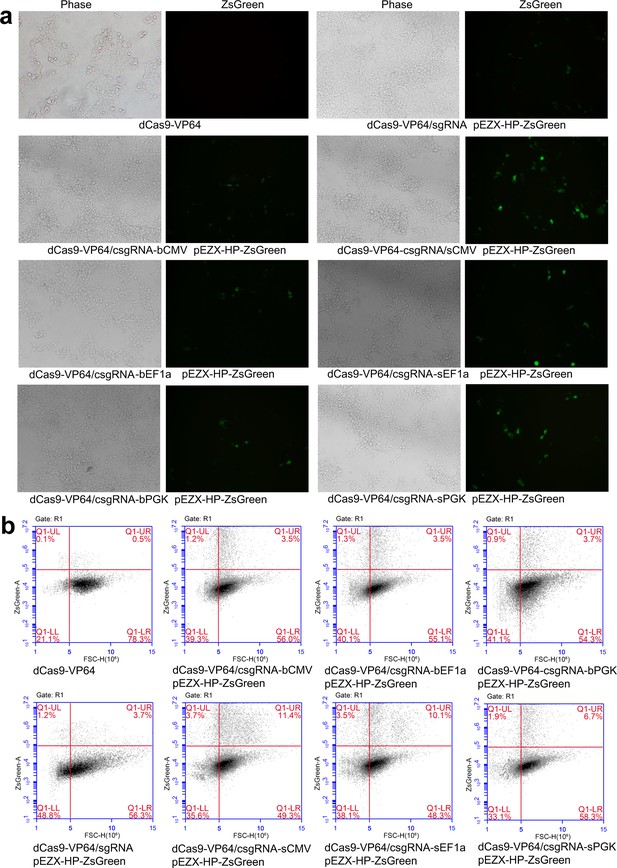
Activation of exogenous and endogenous genes with other trans enhancers.
(a) Activation of exogenous reporter gene ZsGreen. The fluorescence intensity of cells was analyzed with flow cytometry. (b) Activation of endogenous HNF4α gene in 293T and HepG2 cells with two new trans enhancers, sEF1a and sPGK. The sCMV was used as a positive control for comparison. The blunt-end trans enhancers (bEF1a and bPGK) were also used as controls. Data are shown as mean ± SD, n = 3. The statistical difference was analyzed using Student’s t test. *, p<0.05; **, p<0.01.
Activation of exogenous and endogenous genes with other CRISPR-assisted trans enhancers.
(a) Activation of exogenous ZsGreen gene in 293 T cells with two new trans enhancers, sEF1a and sPGK. (b) Flow cytometry analysis of ZsGreen expression.
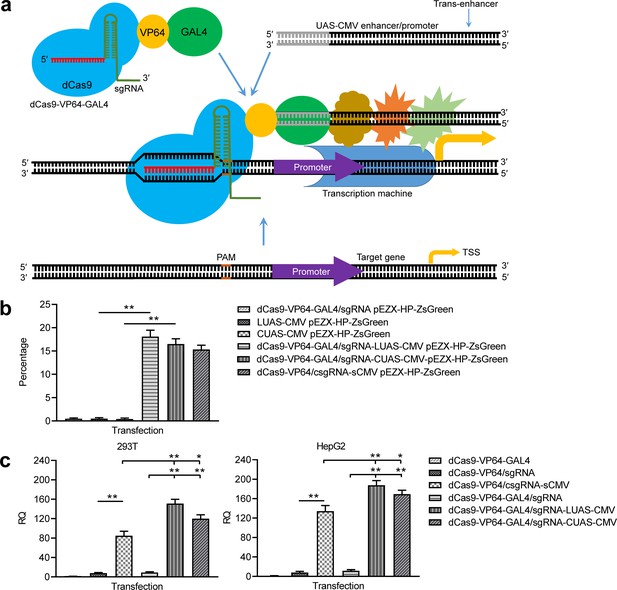
Activation of exogenous and endogenous genes with the GAL4-UAS-based trans enhancer.
(a) Schematic show of gene activation using the GAL4-UAS-based CRISPR-assisted trans enhancer. (b) Activation of exogenous reporter gene ZsGreen. The fluorescence intensity of cells was analyzed with flow cytometry. (c) Activation of endogenous HNF4α gene in 293T and HepG2 cells with the GAL4-UAS-based CRISPR-assisted trans enhancer. The sCMV was used as a positive control for comparison. Three other transfections were used as controls. LUAS-CMV, linear UAS-CMV; CUAS-CMV, circular UAS-CMV. Data are shown as mean ± SD, n = 3. The statistical difference was analyzed by Student’s t-test. *, p<0.05; **, p<0.01.
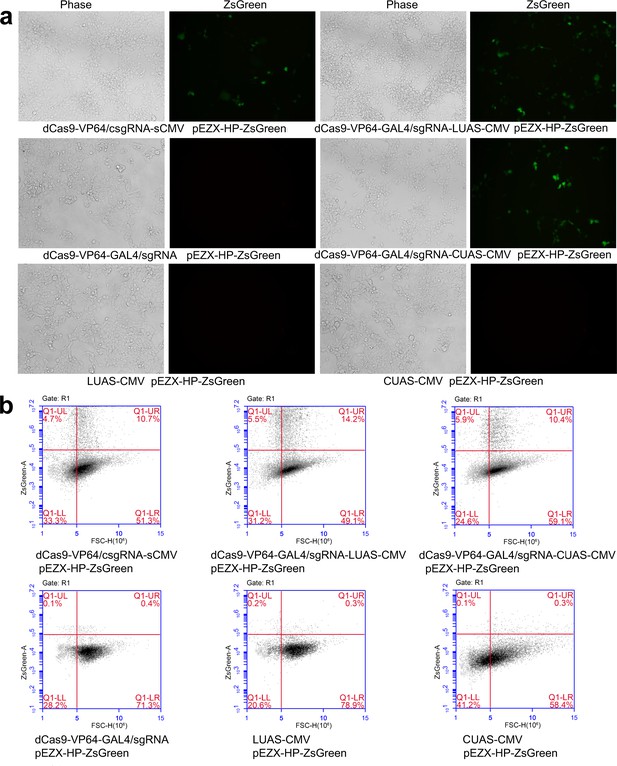
Activation of exogenous and endogenous genes with CRISPR-assisted trans enhancer using the GAL4-UAS system.
(a) Activation of exogenous ZsGreen gene in 293 T cells with the GAL4-UAS-based CRISPR-assisted trans enhancer. (b) Flow cytometry analysis of ZsGreen expression.
Additional files
-
Supplementary file 1
Four tables showing primers and oligos.
- https://doi.org/10.7554/eLife.45973.016
-
Supplementary file 2
Schematic show of construction of sgRNA vectors for blue-white screening.
- https://doi.org/10.7554/eLife.45973.017
-
Supplementary file 3
Sequences of vectors, templates, sgRNA, csgRNA, and trans enhancers.
- https://doi.org/10.7554/eLife.45973.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45973.019





