A positive feedback loop between Myc and aerobic glycolysis sustains tumor growth in a Drosophila tumor model
Figures
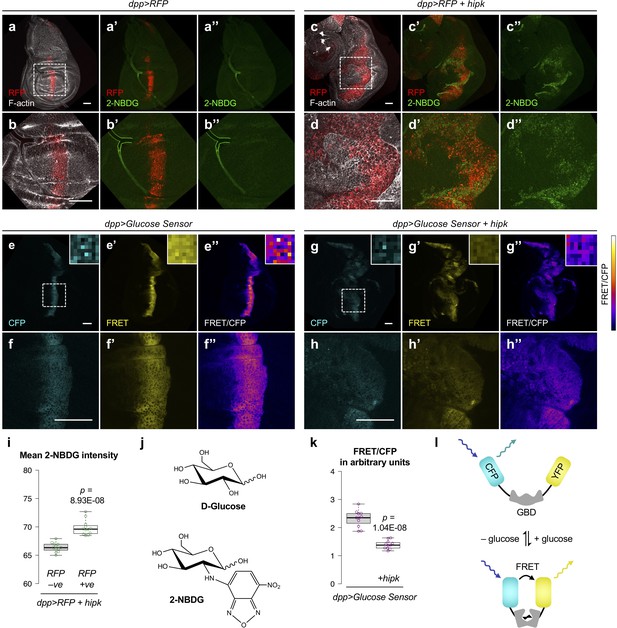
Hipk tumor cells exhibit elevated glucose uptake and reduced intracellular glucose levels.
(a–d) Incorporation of 2-NBDG (green) in control (dpp>RFP) (a, b) and hipk-expressing (dpp>RFP + hipk) wing discs (c, d). Insets (dashed line) in (a and c) are magnified in (b and d), respectively. RFP (red) marks the transgene-expressing cells. F-actin (gray) staining shows the overall tissue morphology. (e–h) Intracellular glucose FRET efficiency (FRET/CFP) (colored using Fire Lookup Table) in control (dpp>Glucose Sensor) (e–f) and hipk-expressing (dpp>Glucose Sensor+hipk) wing discs (g–h). Individual CFP and FRET channels are shown in cyan and yellow, respectively. Insets (dashed line) in (e and g) are magnified in (f and h), respectively. Insets (solid line) at the upper right corners in (e–e’’) and (g–g’’) show the corresponding signals at single pixel level. (i) Quantification of mean 2-NBDG intensity in hipk-expressing cells (RFP positive) and the neighboring wild-type cells (RFP negative) in hipk-expressing wing discs (dpp >RFP + hipk) (n = 12 wing discs). (j) Chemical structures of D-glucose (top) and 2-NBDG (bottom). (k) Quantification of glucose FRET efficiency in control (dpp>Glucose Sensor) and hipk-expressing (dpp >Glucose Sensor + hipk) wing discs (n = 11 wing discs per genotype). (l) Schematic diagram of the FRET-based glucose sensor. Glucose binding induces conformational changes of glucose-binding domain (GBD) such that FRET occurs and YFP emission increases. Scale bars, 50 μm. Exact p values are shown and calculated using unpaired two-tailed t-test.
-
Figure 1—source data 1
Analyses of glucose metabolism in Hipk tumor cells and evaluation of RNAi tools.
- https://doi.org/10.7554/eLife.46315.006
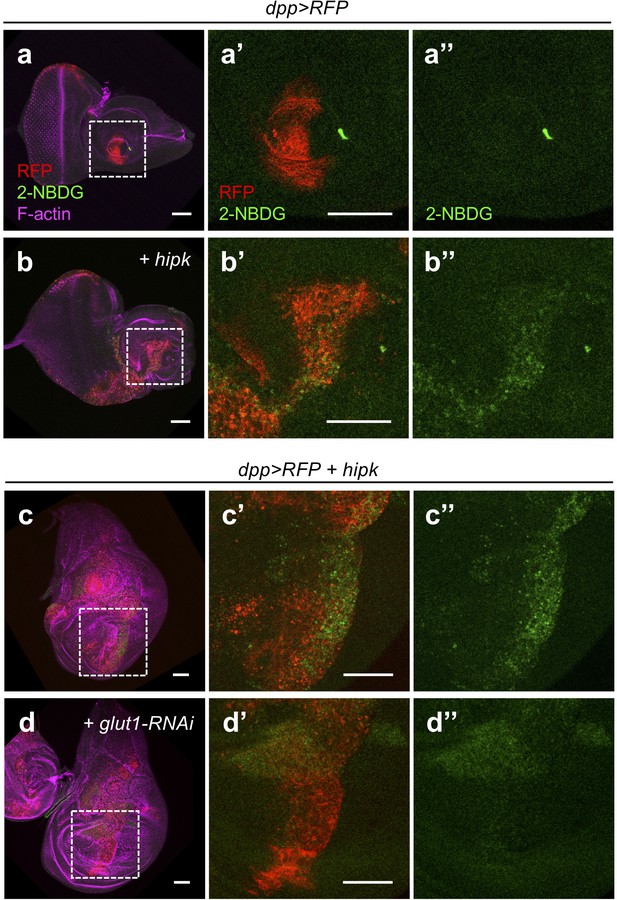
Elevated Hipk stimulates 2-NBDG uptake in a Glut1-dependent manner.
(a–d) Incorporation of 2-NBDG (green) in control (dpp>RFP) (a) and hipk-expressing eye-antennal discs (dpp>RFP + hipk) (b). Insets (dashed line) in (a and b) are magnified in (a’–a’’ and b’–b’’), respectively. RFP (red) marks the transgene-expressing cells. F-actin (magenta) staining shows the overall tissue morphology. (c–d) Incorporation of 2-NBDG (green) in hipk-expressing wing discs without (dpp>RFP + hipk) (c) or with co-expression of glut1-RNAi (dpp>RFP + hipk + glut1-RNAi) (d). Insets (dashed line) in (c and d) are magnified in (c’–c’’ and d’–d’’), respectively. RFP (red) marks the transgene-expressing cells. F-actin (magenta) staining shows the overall tissue morphology. Scale bars, 50 μm.
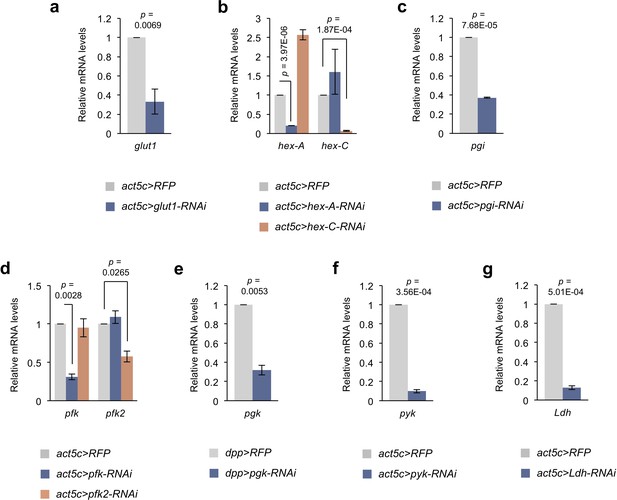
Knockdown efficiencies of the RNAi lines targeting glycolytic genes.
(a) qRT-PCR analysis of glut1 expression in control (act5c>RFP) and glut1 knockdown (act5c>glut1-RNAi) pupae. (b) qRT-PCR analyses of hex-A and hex-C expression in control (act5c>RFP), hex-A knockdown (act5c>hex A-RNAi) and hex-C knockdown (act5c>hex C-RNAi) pupae. (c) qRT-PCR analysis of pgi expression in control (act5c>RFP) and pgi knockdown (act5c>pgi-RNAi) pupae. (d) qRT-PCR analyses of pfk and pfk2 expression in control (act5c>RFP), pfk knockdown (act5c>pfk-RNAi) and pfk2 knockdown (act5c>pfk2-RNAi) larvae. (e) qRT-PCR analysis of pgk expression in control (dpp>RFP) and pgk knockdown (dpp>pgk-RNAi) larval salivary gland cells. (f) qRT-PCR analysis of pyk expression in control (act5c>RFP) and pyk knockdown (act5c>pyk-RNAi) larvae. (g) qRT-PCR analysis of Ldh expression in control (act5c>RFP) and Ldh knockdown (act5c>Ldh-RNAi) pupae. Data are mean ± sem. At least n = 2 independent experiments with samples pooled from approximately 10 larvae or pupae. Exact p values are shown and calculated using unpaired two-tailed t-test.
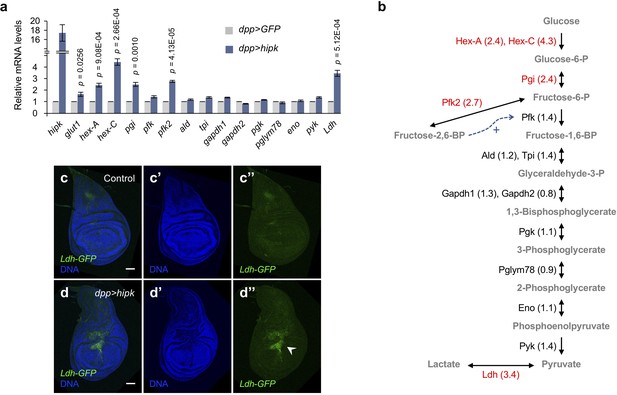
Elevated Hipk induces robust expression of a subset of glycolytic genes.
(a, b) qRT-PCR analysis of glycolytic gene expression in hipk-expressing discs (dpp>hipk) relative to control discs (dpp>GFP) (a). Data are mean ± sem. n = 3 independent experiments with samples pooled from 10 to 20 wing discs per genotype. Schematic diagram of glycolysis (b). Fold changes are shown in brackets. Glycolytic enzymes with more than 1.5-fold transcriptional upregulation in hipk-expressing discs are marked in red. Bi- and uni-directional arrows indicate the reversible and essentially irreversible steps in glycolysis, respectively. Dashed arrow (blue) shows the positive allosteric regulation of Pfk by fructose-2,6-BP. Full names of the glycolytic genes are provided in Supplementary file 1. Exact p values are shown and calculated using unpaired two-tailed t-test. (c–d) Expression of Ldh-GFP (green), an enhancer trap to monitor Ldh expression, in control (genotype: Ldh-GFP/+) (c) and hipk-expressing wing discs (genotype: Ldh-GFP/dpp>hipk) (d). DAPI staining for DNA (blue) reveals tissue morphology. White arrowhead in d’’ indicates marked induction of Ldh expression. Scale bars, 50 μm.
-
Figure 2—source data 1
Expression profile of glycolytic genes inhipk-expressing discs.
- https://doi.org/10.7554/eLife.46315.008
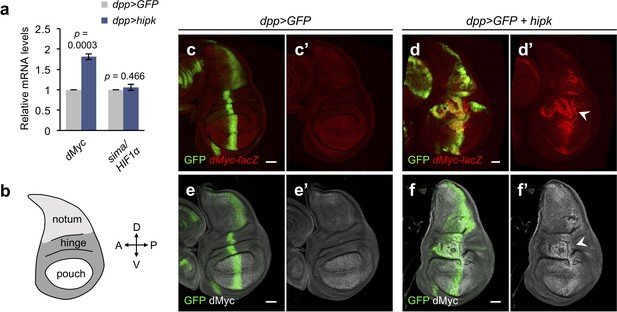
Elevated Hipk drives dMyc upregulation in a region-specific manner.
(a) qRT-PCR analysis of dMyc and sima expression in hipk-expressing discs (dpp>hipk) relative to control discs (dpp>GFP). Data are mean ± sem. n = 3 independent experiments with samples pooled from 10 to 20 wing discs per genotype. Exact p values are shown and calculated using unpaired two-tailed t-test. (b) Schematic diagram of a larval wing imaginal disc, with notum, hinge and pouch in light gray, dark gray and white, respectively. (c–d) β-galactosidase staining (red) in control (dpp>GFP) (c) and hipk-expressing (dpp>GFP + hipk) (d) wing discs harboring a lacZ reporter to monitor the transcriptional induction of dMyc (dMyc-lacZ). White arrowhead in (d’) indicates robust upregulation of dMyc in the wing hinge. (e–f) dMyc staining (gray) in control (dpp>GFP) (e) and hipk-expressing (dpp>GFP + hipk) (f) wing discs. White arrowhead in (f’) indicates dMyc accumulation in the wing hinge. GFP (green) marks the transgene-expressing cells. Scale bars, 50 μm.
-
Figure 3—source data 1
Expression levels ofdMycandsimainhipk-expressing discs.
- https://doi.org/10.7554/eLife.46315.010
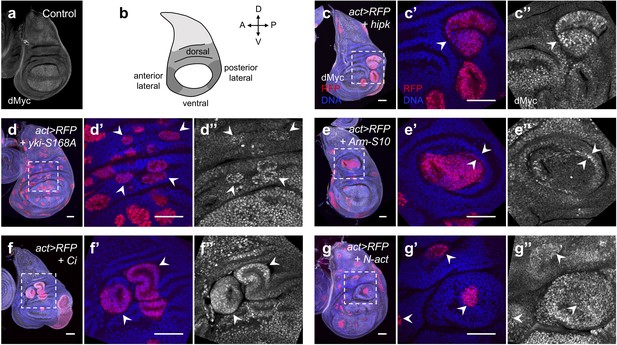
Elevated Hipk and perturbations in cell signaling cascades induce dMyc accumulation in the wing hinge region.
(a) dMyc staining (gray) in a control wing disc. (b) Schematic diagram of a larval wing imaginal disc. The hinge region can be divided into dorsal, lateral and ventral compartments. (c–g) dMyc staining (gray) in flip-out clones expressing hipk (c), yki-S168A (d), Arm-S10 (e), Ci (f) or N-act (g) under the control of actin-Gal4. RFP (red) marks the transgene-expressing clones. DAPI staining for DNA (blue) reveals tissue morphology. (c’, c’’-g’, g’’) are magnified images of the insets (dashed lines) in (c-g). (c) White arrowhead indicates dMyc accumulation in a hipk-expressing clone. (d) White arrowheads indicate yki-S168A-expressing clones with dMyc accumulation. (e) White arrowheads indicate ectopic dMyc expression at the boundary of the Arm-S10-expressing clone and the neighboring wild-type cells. (f) White arrowheads indicate dMyc upregulation in Ci-expressing clones. (g) White arrowheads indicate dMyc accumulation in N-act-expressing clones and the adjacent wild-type cells. Scale bars, 50 μm.
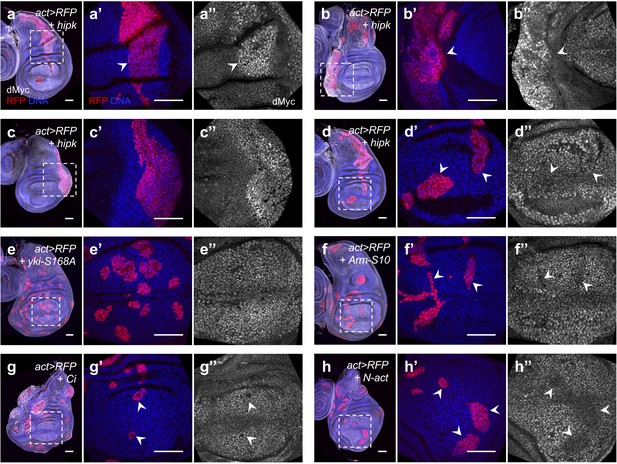
Effects on dMyc expression by elevated Hipk or perturbations in cell signaling.
(a–h) dMyc staining (gray) in flip-out clones expressing hipk (a–d), yki-S168A (e), Arm-S10 (f), Ci (g) or N-act (h) under the control of actin-Gal4. RFP (red) marks the transgene-expressing clones. DAPI staining for DNA (blue) reveals tissue morphology. (a’, a’’-h’, h’’) are magnified images of the insets (dashed lines) in (a–h). Scale bars, 50 μm.
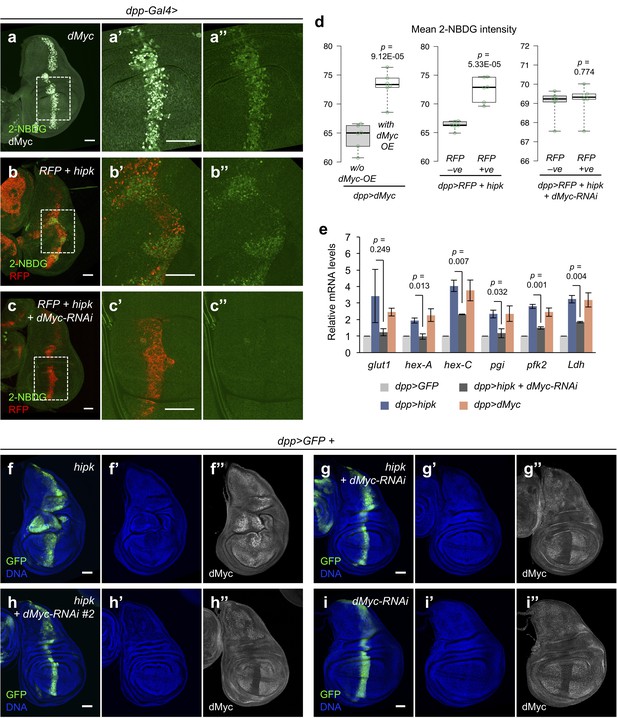
dMyc mediates the Hipk-induced glycolysis and tumor growth.
(a–c) Incorporation of 2-NBDG (green) in dMyc-expressing wing disc (dpp>dMyc) (a), hipk-expressing wing disc (dpp>RFP + hipk) (b), and hipk-expressing wing disc with dMyc knockdown (dpp>RFP + hipk + dMyc-RNAi) (c). Insets (dashed line) in (a, b and c) are magnified in (a’–a’’), (b’–b’’) and (c’–c’’), respectively. dMyc staining (gray) in (a) marks the dMyc-overexpressing cells. RFP (red) in (b and c) marks the transgene-expressing cells. (d) Quantification of mean 2-NBDG intensity in dMyc-expressing cells and the neighboring wild-type cells in dMyc-expressing wing discs (dpp>dMyc) (left), in hipk-expressing cells (RFP positive) and the neighboring wild-type cells (RFP negative) in hipk-expressing discs (dpp>RFP + hipk) (middle) and, in hipk, dMyc-RNAi co-expressing cells (RFP positive) and the neighboring wild-type cells (RFP negative) in hipk-expressing discs with dMyc knockdown (dpp>RFP + hipk + dMyc-RNAi) (right). n = 6 discs per genotype. (e) qRT-PCR analysis of expression of a set of glycolytic genes in hipk-expressing wing discs (dpp>hipk), hipk-expressing discs with dMyc knockdown (dpp>hipk + dMyc-RNAi), and dMyc-expressing discs relative to control discs (dpp>GFP). Data are mean ± sem. n = 3 independent experiments with samples pooled from 10 to 20 wing discs per genotype. (f–i) dMyc staining (gray in f’’), (g’’, h’’ and i’’) in hipk-expressing disc (dpp>GFP + hipk) (f), hipk and dMyc-RNAi co-expressing disc (dpp>GFP + hipk + dMyc-RNAi) (g), hipk and dMyc-RNAi #2 co-expressing disc (dpp>GFP + hipk + dMyc-RNAi #2) (h) and dMyc knockdown disc (dpp>GFP + dMyc-RNAi) (i). GFP (green) marks the transgene-expressing cells. DAPI staining for DNA (blue) reveals tissue morphology. Scale bars, 50 μm. Exact p values are shown and calculated using unpaired two-tailed t-test.
-
Figure 5—source data 1
Analyses of glucose metabolism inhipk-expressing discs upondMycknockdown.
- https://doi.org/10.7554/eLife.46315.014
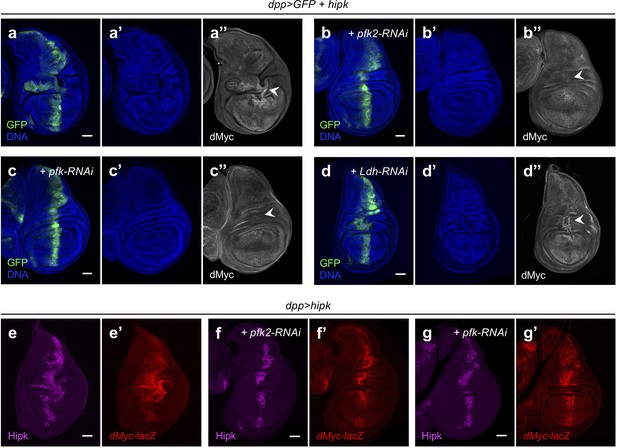
Pfk2 and Pfk sustain Hipk tumor growth through post-transcriptional regulation of dMyc.
(a–d) dMyc staining (gray in a’’, b’’, c’’ and d’’) in hipk-expressing wing disc (dpp>GFP + hipk) (a), hipk and pfk2-RNAi co-expressing disc (dpp>GFP + hipk + pfk2-RNAi) (b), hipk and pfk-RNAi co-expressing disc (dpp>GFP + hipk + pfk-RNAi) (c) and hipk and Ldh-RNAi co-expressing disc (dpp>GFP + hipk + Ldh-RNAi) (d). GFP (green) marks the transgene-expressing cells. DAPI staining for DNA (blue) reveals tissue morphology. (e–g) Hipk staining (magenta in e, f and g) and β-galactosidase staining (red in e’, f’ and g’) in hipk-expressing disc (dpp>hipk) (e), hipk and pfk2-RNAi co-expressing disc (dpp>hipk + pfk2-RNAi) (f), hipk and pfk-RNAi co-expressing disc (dpp>hipk + pfk-RNAi) (g). All wing discs harbor a lacZ reporter to monitor the transcriptional induction of dMyc (dMyc-lacZ). Scale bars, 50 μm.
-
Figure 6—source data 1
Data for pyruvate measurements.
- https://doi.org/10.7554/eLife.46315.021
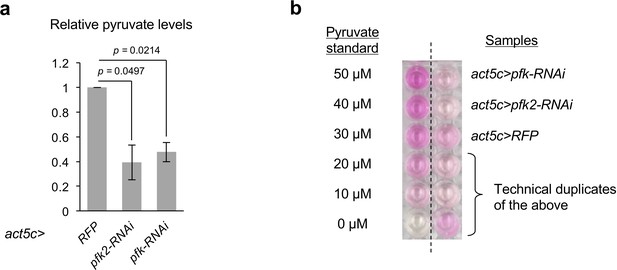
Larvae depleted of pfk2 or pfk display decreases in the pyruvate levels.
(a) Relative pyruvate levels in control (act5c>RFP), pfk2 knockdown (act5c>pfk2-RNAi) and pfk knockdown (act5c>pfk-RNAi) larvae. Data are mean ± sem and pooled from two biological replicates. Exact p values are shown and calculated using unpaired two-tailed t-test. (b) The representative result of the colorimetric assay for pyruvate measurement from one set of biological samples. The left column shows the pyruvate standards. The right column shows the biological samples with the indicated genotypes.
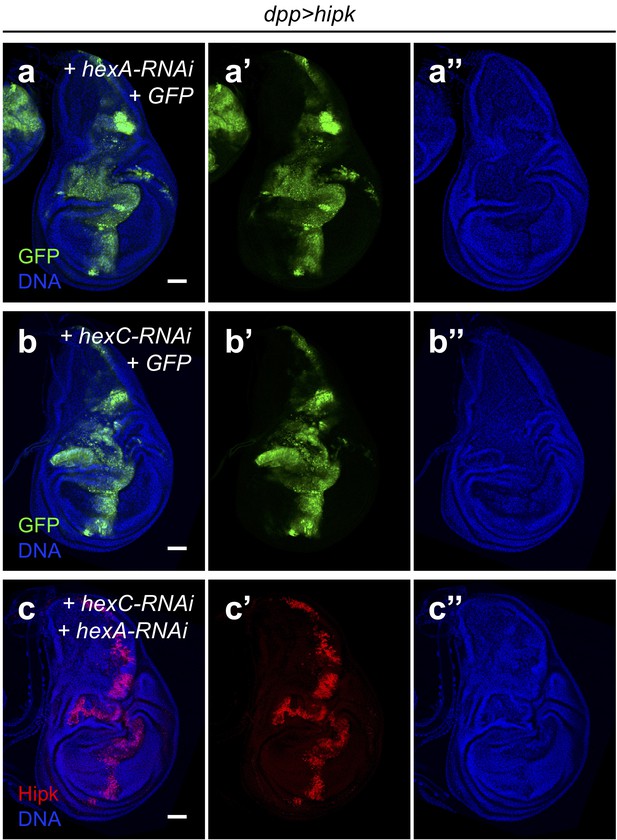
Hexokinases are not required for Hipk tumor growth.
(a–b) hipk-expressing wing disc with hex-A knockdown (a) or hex-C knockdown (b). GFP (green) marks the transgene-expressing cells. (c) hipk-expressing wing disc with hex-A and hex-C double knockdown. Hipk staining (red) marks the transgene-expressing cells. DAPI staining for DNA (blue) reveals tissue morphology. Scale bars, 50 μm.
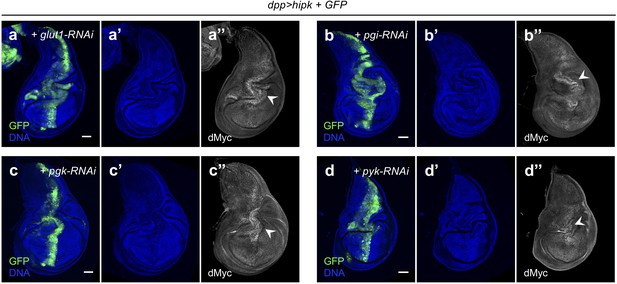
Non-targeted inhibition of glycolysis is not sufficient to prevent Hipk tumor growth or ectopic dMyc accumulation.
(a–d) dMyc staining (gray in a’’), (b’’, c’’ and d’’) in hipk and glut1-RNAi co-expressing disc (dpp>GFP + hipk + glut1-RNAi) (a), hipk and pgi-RNAi co-expressing disc (dpp>GFP + hipk + pgi-RNAi) (b), hipk and pgk-RNAi co-expressing disc (dpp>GFP + hipk + pgk-RNAi) (c) and hipk and pyk-RNAi co-expressing disc (dpp>GFP + hipk + pyk-RNAi) (d). GFP (green) marks the transgene-expressing cells. DAPI staining for DNA (blue) reveals tissue morphology. Scale bars, 50 μm.
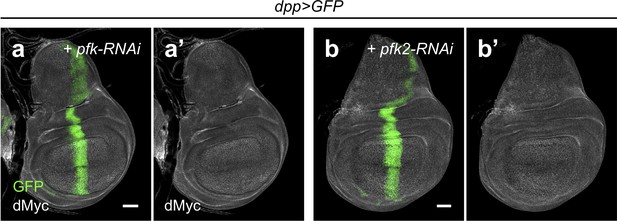
Pfk2 and Pfk do not affect dMyc expression in the absence of hipk overexpression.
(a–b) dMyc staining (gray) in pfk knockdown discs (dpp>GFP + pfk-RNAi) (a) and pfk2 knockdown discs (dpp>GFP + pfk2-RNAi) (b). GFP (green) marks the transgene-expressing cells. Scale bars, 50 μm.
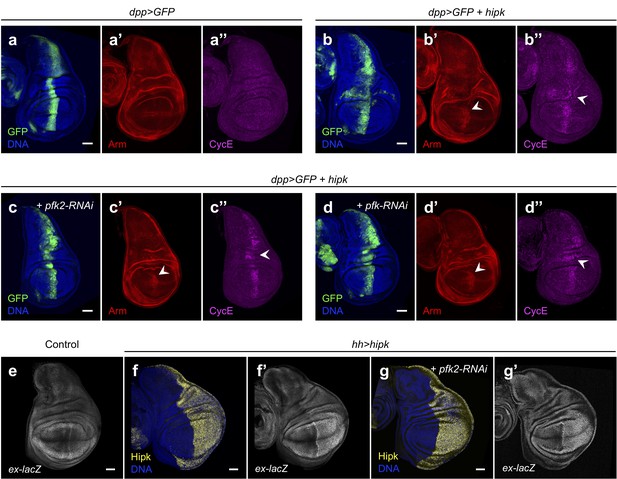
Knockdown of pfk2 or pfk does not affect Hipk functions.
(a–b) Armadillo (Arm, red) and Cyclin E (CycE, magenta) in control discs (dpp>GFP) (a), hipk-expressing discs (dpp>GFP + hipk) (b), hipk-expressing discs with pfk2 knockdown (dpp>GFP + hipk + pfk2-RNAi) (c), hipk-expressing discs with pfk knockdown (dpp>GFP + hipk + pfk-RNAi) (d). White arrowheads in (b’, c’ and d’) indicate stabilization of Arm in the pouch region. White arrowheads in (b’’, c’’ and d’’) indicate CycE induction along the anterior/posterior boundary of the wing discs. GFP (green) marks the transgene-expressing cells. (e–g) β-galactosidase staining (gray) in control disc (hh-Gal4) (e), hipk-expressing disc (hh>hipk) (f) and hipk-expressing disc with pfk2 knockdown (hh>hipk + pfk2-RNAi) (g). Hh-Gal4 drives transgenic expression in the posterior compartment of wing discs (left half of the discs). All wing discs harboring a lacZ reporter to monitor the transcriptional induction of expanded (ex-lacZ). Hipk staining (yellow) in (f and g) marks the transgene-expressing cells (posterior compartment). Scale bars, 50 μm.
Additional files
-
Supplementary file 1
A list of Drosophila genes studied in this work with their human homologs.
- https://doi.org/10.7554/eLife.46315.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46315.023





