HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations
Figures
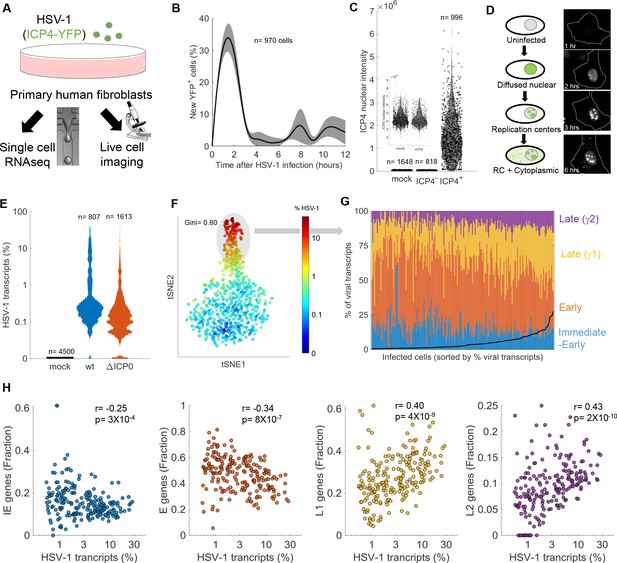
Cell-to-cell variability in infection dynamics and viral gene expression.
(A) HDFn cells were infected with HSV-1 expressing ICP4-YFP and analyzed by time-lapse fluorescent imaging and scRNA-seq. (B) Distribution of the initial time of ICP4 expression. The black line denotes the average and the gray shadowing denotes the standard error of the time of ICP4 expression by single cells. n = 970 cells from three fields of view. (C) Violin plots showing the distribution of ICP4 nuclear intensity at 5 hr post infection of mock (n = 1648) or HSV-1 infected cells, either ICP4– (n = 818) or ICP4+ (n = 996). The smaller inset shows a zoomed-in view of the mock and ICP4– cells. Values of single cells are shown as circles. (D) Schematic diagram of the different localization phenotypes of ICP4 (left) and a representative single cell showing these phenotypes at different time points following infection. (E) Violin plots showing the distribution of the % of viral transcripts (out of the total transcripts) for individual cells that were mock-infected (n = 4500), infected with wt (n = 807) or infected with ΔICP0 (n = 1613) HSV-1. The y-axis is logarithmic. (F) T-distributed stochastic neighbor embedding (tSNE) plot based on viral gene expression in wt HSV-1 infection. Each dot represents a single cell and is colored according to the % of viral transcripts from blue (low) to red (high). The color bar is logarithmic. Gini is the Gini coefficient. (G) The relative abundance of the four viral gene classes: immediate-early (blue), early (orange), late γ1 (yellow) and late γ2 (purple) in highly infected single cells (highlighted by the gray oval in panel (F)). Single cells are ordered by their % of viral transcripts from low (left) to high (right), which are denoted by the black line. (H) Scatter plots of single cells showing the % of viral transcripts on the x-axis and the relative abundance of each viral gene class on the Y-axis. r and p are the Pearson correlation coefficients and p-values, respectively.
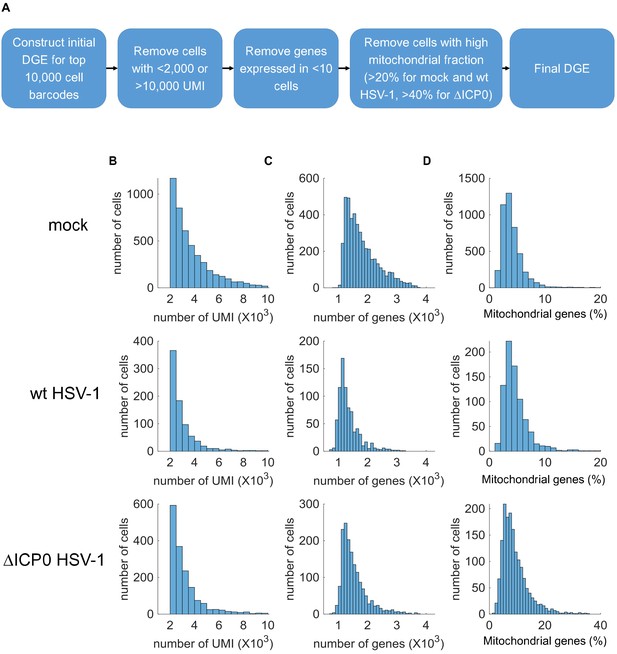
Technical data relating to single-cell RNA-sequencing.
(A) Flow chart describing the generation of a DGE (digital expression matrix) from raw sequencing data and the filtering criteria applied at different stages to the generation of the final DGE. Note that the threshold for mitochondrial gene expression is different for ΔICP0-infected cells (>40% rather than 20% for mock and wt-infected), because ΔICP0 infection caused an increase in mitochondrial gene expression. (B) Distribution of read depth (number of unique molecular identifiers (# UMI)) per cell. (C) Distribution of read depth (# genes) per cell. (D) Distribution of mitochondrial gene expression (note that the scale of the x-axis is different for ΔICP0).
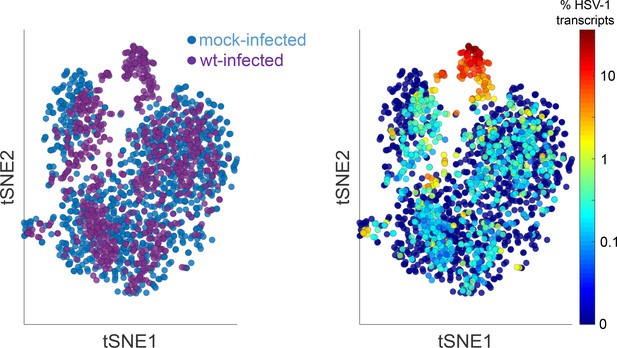
Joint analysis of host +viral genes in mock and wt-infected HDFn.
The left panel shows the identity of the cells (blue = mock, purple = wt infected) and the right panel shows the level of HSV-1 gene expression (blue = low, red = high). Highly infected cells cluster separately from the mock-infected and low-infected cells, which are intermingled.
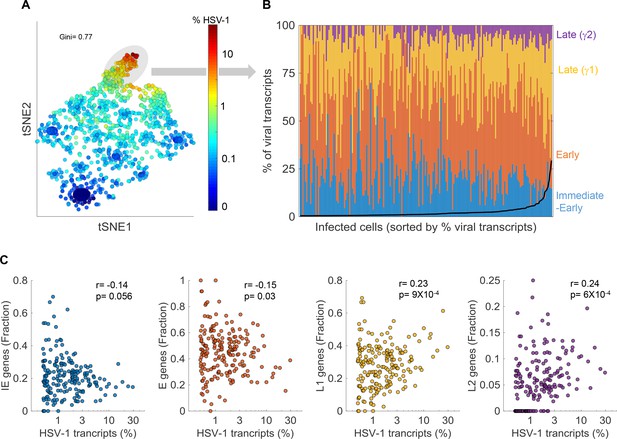
Cell-to-cell variability in viral gene expression upon ΔICP0 infection.
(A) tSNE plot based on viral gene expression. Each dot represents a single cell and is colored according to the % of viral transcripts from blue (low) to red (high). The color bar is logarithmic. Gini is the Gini coefficient. (B) The relative abundance of the four viral gene classes: immediate-early (blue), early (orange), late γ1 (yellow) and late γ2 (purple) in highly infected single cells (highlighted by the gray oval in panel (A)). Single cells are ordered by their % of viral transcripts from low (left) to high (right), which are denoted by the black line. (C) Scatter plots of single cells showing the % of viral transcripts on the x-axis and the relative abundance of each viral gene class on the y-axis. r and p are the Pearson correlation coefficients and p-values, respectively.
Live imaging of HDFn infected by wildtype HSV-1 expressing ICP4-YFP.
https://doi.org/10.7554/eLife.46339.007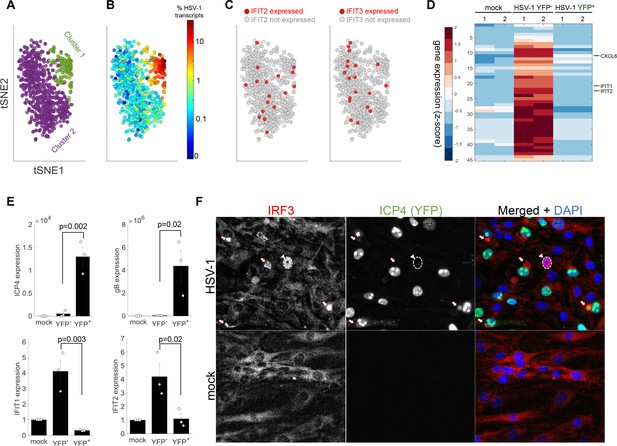
The anti-viral program is initiated in a rare sub-population of abortively infected cells.
(A) tSNE plot based on viral and host gene expression. Cells are colored according to their clustering. Cluster one is colored green and cluster two is colored purple. (B) tSNE plot as in panel (A). Cells are colored according to their % of viral transcript expression. (C) tSNE plot as in panel (A). Cells are colored according to their expression (red) or lack of expression (gray) of IFIT2 (left) or IFIT3 (right). (D) Heat-map of genes that are significantly upregulated in ICP4– cells, as compared to both mock and ICP4+ cells. RNA-sequencing was performed in duplicates denoted by the numbers 1 and 2 on the top row. Each row shows the normalized (z-score) expression of a single gene, colored from low (blue) to high (red). (E) Validation of selected genes by quantitative PCR. Bar plots show the expression levels of the viral genes ICP4 and gB (top) and the anti-viral genes IFIT1 and IFIT2 (bottom). Values are mean ± s.e. of three independent biological repeats. Individual measurements are shown as circles. p-values were calculated using a one-tailed t-test. (F) Immunoflorescent staining of IRF3 in mock-infected (bottom) or HSV-1 infected (top) cells at 5 hr post infection. The arrowhead points to an ICP4-negative cell that shows nuclear IRF3 staining (nucleus border denoted by a dashed white line). The arrows point to aggregations of IRF3 outside the nucleus in ICP4-positive cells.
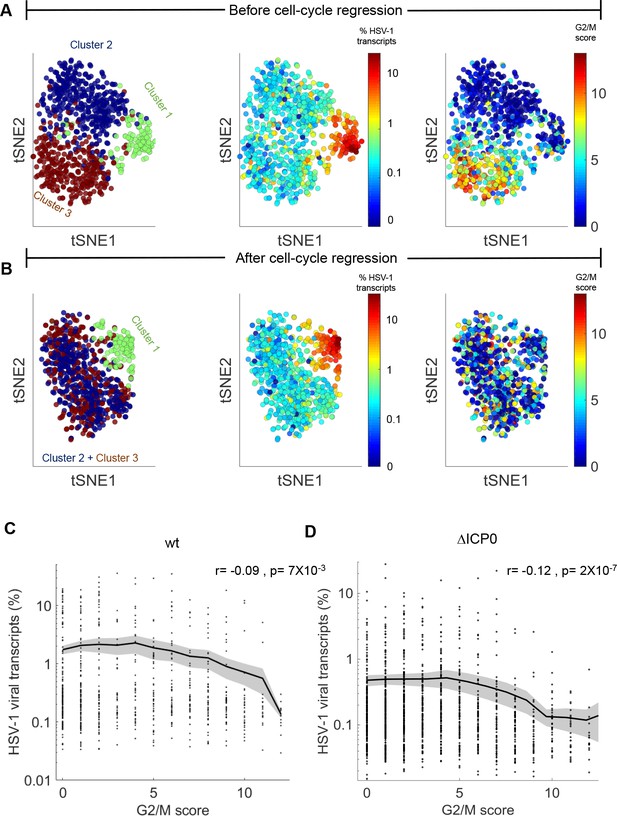
Cell-cycle is anti-correlated with viral gene expression and is a major source of transcriptional variability.
(A) tSNE plots based on viral and host gene expression (wildtype infection). (Left) Cells are colored according to their clustering. Cluster one is colored green, cluster two is colored blue and cluster three is colored brown. (Middle) Cells are colored on the basis of the % of viral transcripts they express from blue (low) to red (high). The color bar is logarithmic. (Right) Cells are colored on the basis of their cell-cycle score, from blue (low) to red (high). (B) tSNE plots as in panel (A), showing cell clustering (left), viral gene expression (middle) and cell-cycle score (right) after regressing out the cell-cycle effect on gene expression. (C,D) Cell-cycle score is anti-correlated with wildtype (C) and ΔICP0 (D) viral transcription. r and p are the Pearson correlation coefficient and the associated p-value, respectively.
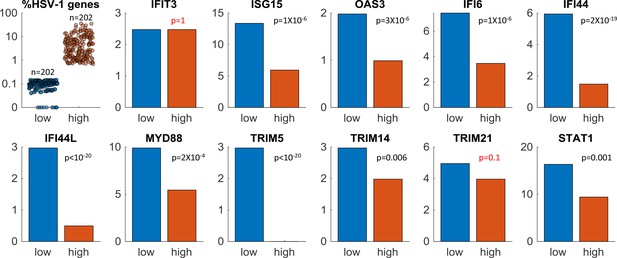
ISGs expression in single-cells infected by wildtype HSV-1.
Here we compare the expression of interferon-stimulated genes (ISGs) in the top (orange) and bottom (blue) 25% of cells, sorted according to the level of HSV-1 gene expression. The top left panel shows the % expression of HSV-1 genes in individual cells in each group (n = 202 cells in each). The remaining panels show the % of cells that express a given ISG in each group. p-values were calculated using the Chi-square test and false discovery rate (FDR)-corrected for multiple comparisons. Note that two ISGs (IFIT3 and TRIM21) have p-values >0.05.
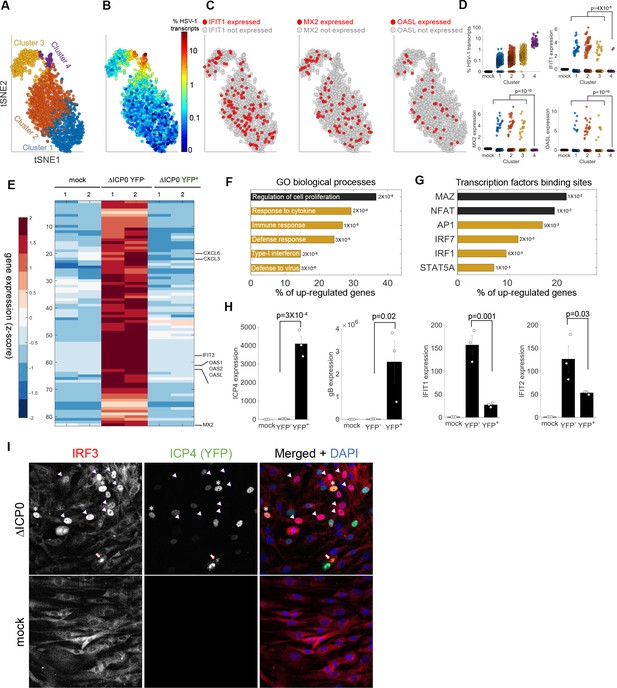
Anti-viral signaling in cells infected by ΔICP0 mutant.
(A) tSNE plot based on viral and host gene expression. Cells are colored according to their clustering. (B) tSNE plot as in panel (A), with cells colored according to the % expressed transcripts that are viral. (C) tSNE plots as in panel (A). Cells are colored according to expression (red) or lack of expression (gray) of IFIT1 (left), MX2 (middle) or OASL (right). (D) Scatter plots showing the expression of HSV-1 transcripts (top left) and the expression of the anti-viral genes IFIT1, MX2 and OASL in mock-infected cells (first column) and in each cluster of cells infected by ΔICP0. p-values were calculated by a Chi-square test, comparing cluster 4 to clusters 1–3. (E) Heat-map of genes that are significantly upregulated in ΔICP0-infected ICP4– cells, as compared to both mock and ICP4+ cells. RNA-sequencing was performed in duplicates, as denoted by the numbers 1 and 2 on the top row. Each row shows the normalized (z-score) expression of a single gene, colored from low (blue) to high (red). (F) Gene Ontology (GO) terms for the enriched genes identified in panel (E). The term is written on the bar, and the value next to each bar is the FDR-corrected p-value for its enrichment. Bar length shows the % of upregulated genes annotated to the GO term. Highlighted in yellow are GO terms that are associated with anti-viral signaling. (G) Transcription factor binding sites that are enriched in the promoters of genes identified in panel (E). (H) QPCR validation of selected genes. The bar plots show the expression level of the viral genes ICP4 and gB and of the anti-viral genes IFIT1 and IFIT2. Values are means ± s.e. of three independent biological repeats. Individual measurements are shown as circles. p-values were calculated using a one-tailed t-test. (I) Immunoflorescent staining of IRF3 in mock-infected (bottom) or ΔICP0-infected (top) cells at 5 hr post infection. The arrowheads point to ICP4-negative cells that contain nuclear IRF3. Asterisks denote ICP4-positive cells that contain nuclear IRF3. The arrow points to a cell in the late stages of infection, in which IRF3 is aggregated in the nuclear periphery.
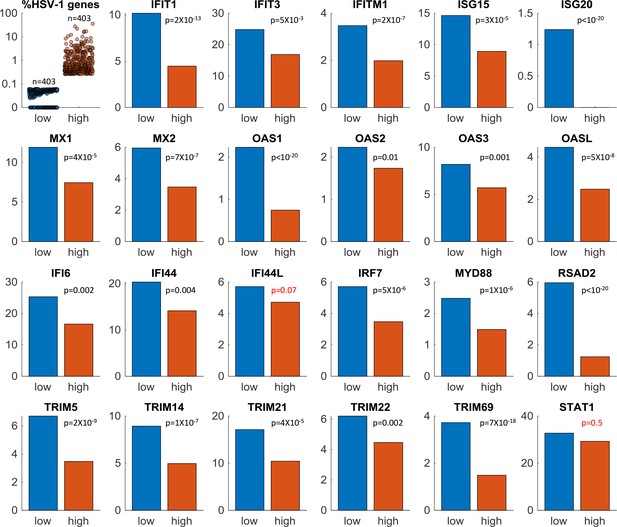
ISGs expression in single-cells infected by ΔICP0 HSV-1.
Here we compare the expression of interferon-stimulated genes) (ISGs) in the top (orange) and bottom (blue) 25% of cells, sorted according to the level of HSV-1 gene expression. The top left panel shows the % expression of HSV-1 genes in individual cells in each group (n = 403 cells in each). The remaining panels show the % of cells expressing a given ISG in each group. p-values were calculated using the Chi-square test and FDR-corrected for multiple comparisons. Note that two ISGs (IFI44L and STAT1) have p-values >0.05.

HSV-1 infection upregulates developmental pathways.
(A) Heat-map of genes that are significantly upregulated in ICP4+ cells, as compared to both mock-infected and ICP4– cells. RNA-sequencing was performed in duplicates denoted by the numbers 1 and 2 on the top row. Each row shows the normalized (z-score) expression of a single gene, colored from low expression (blue) to high expression (red). (B) Heat-map of genes that are significantly upregulated in cluster 1 (highly infected) compared to cluster 2 (lowly infected) single cells. Each row shows the normalized (z-score) expression of a single gene, colored from low expression (blue) to high expression (red) in 80 single cells (40 from cluster 1 and 40 from cluster 2). The color bar is shared for both panels (A) and (B). (C,D) GO terms enriched for genes identified in panels (A) and (B), respectively. The GO term is written on the bar and the value next to each bar is the FDR-corrected p-value for its enrichment. Bar length shows the % of upregulated genes annotated to the GO term. GO terms that are associated with development and gene regulation are highlighted in yellow. (E,F) Transcription-factor-binding sites that are enriched in the promoters of genes identified in panels (A) and (B), respectively. The value next to each bar is the FDR-corrected p-value for the enrichment of the corresponding transcription-factor-binding site. Bar length shows the % of upregulated genes that contain binding sites for each transcription factor. Highlighted in yellow are the LEF1, TCF3, MYC and NMYC transcription factors, which are part of the WNT/β-catenin pathway. (G) A simplified diagram of the WNT/ β-catenin signaling pathway that controls LEF/TCF transcriptional activity.
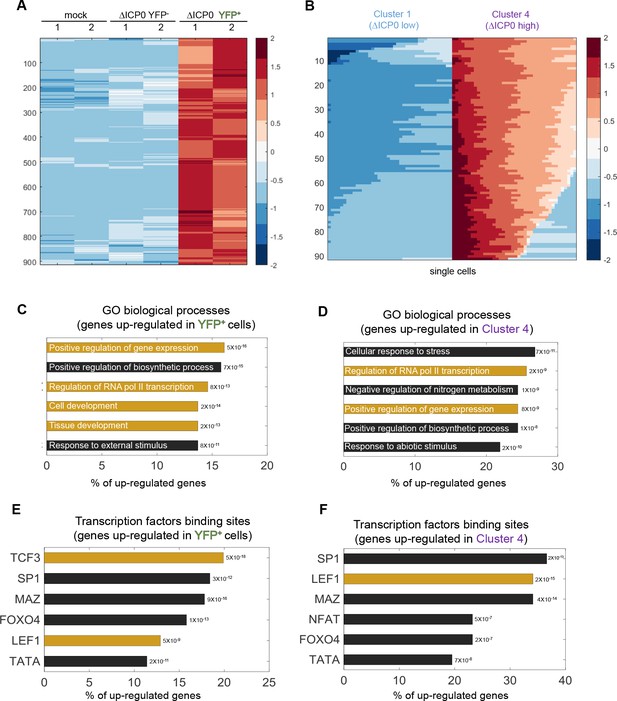
ΔICP0 infection upregulates developmental pathways.
(A) Heat-map of genes that are significantly upregulated in ICP4+ cells, as compared to both mock-infected and ICP4– cells. RNA-sequencing was performed in duplicates denoted by the numbers 1 and 2 on the top row. Each row shows the normalized (z-score) expression of a single gene, colored from low expression (blue) to high expression (red). (B) Heat-map of genes that are significantly upregulated in cluster 4 (highly infected) compared to cluster 1 (lowly infected) single cells. Each row shows the normalized (z-score) expression of a single gene, colored from low expression (blue) to high expression (red) in 80 single cells (40 from cluster 4 and 40 from cluster 1). (C,D) GO terms enriched for genes identified in panels (A) and (B), respectively. The GO term is written on the bar and the value next to each bar is the FDR-corrected p-value for its enrichment. Bar length shows the % of upregulated genes annotated to the GO term. GO terms associated with development and gene regulation are highlighted in yellow. (E,F) Transcription-factor-binding sites that are enriched in the promoters of genes identified in panels (A) and (B), respectively. The value next to each bar is the FDR-corrected p-value for the enrichment of the corresponding transcription-factor-binding site. Bar length shows the % of upregulated genes that contain binding sites for each transcription factor. Highlighted in yellow are the LEF1 and TCF3 transcription factors, which are part of the WNT/β-catenin pathway.
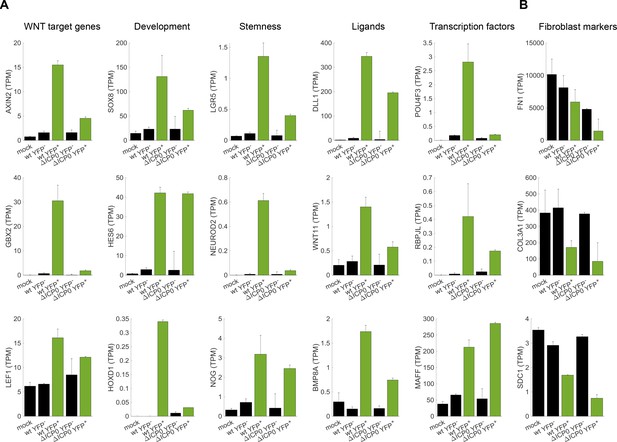
Cellular reprogramming during HSV-1 infection.
(A) Bar plots showing the expression level (transcripts per million (TPM)) of selected examples of genes that participate in developmental pathways and are upregulated in HSV-1-infected cells. Black bars denote mock-infected and ICP4– cells. Green bars denote ICP4+ cells. Values are means ±s.e of the two sequenced biological replicates (Figure 4A and Figure 4—figure supplement 1A). (B) Bar plots showing the expression level of selected examples of fibroblast marker genes.
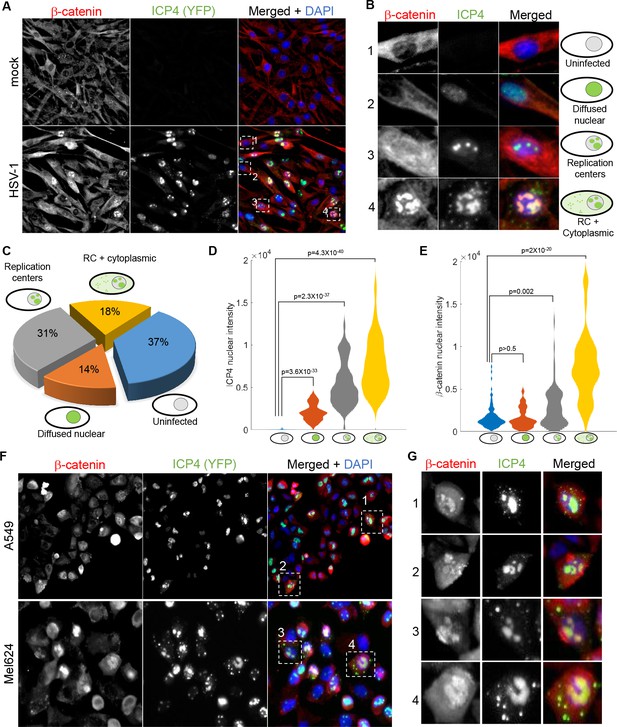
β-catenin translocates to the nucleus and concentrates in the viral replication compartments.
(A) Immunoflorescent staining of β-catenin in mock-infected (top) or HSV-1 infected (bottom) HDFn cells at 5 hr post infection. (B) Magnified images of the four cells denoted by dashed white boxes in panel (A), showing representative images of cells with different ICP4 and β-catenin localizations. (C) Quantification of the relative abundances of the different ICP4 localizations (n = 204 cells). (D,E) Violin plots showing the distributions of ICP4 (D) and β-catenin (E) nuclear levels in cells showing the four ICP4 localization phenotypes. p-values were calculated by a two-tailed two-sample t-test and corrected for multiple comparisons by the Bonferroni correction. (F,G) Immunofluorsence as in panels (A) and (B) for A549 and Mel624 cells.
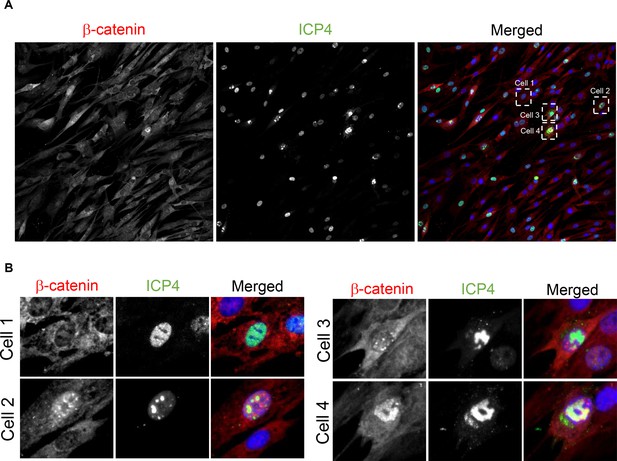
β-catenin translocates to the nucleus and concentrates in the viral replication compartments upon ΔICP0 infection.
(A) Immunofluorescent staining of β-catenin in ΔICP0-infected HDFn cells at 5 hr post infection. (B) Magnified images of the four cells denoted by dashed white boxes in panel (A), showing representative images of cells with different ICP4 and β-catenin localizations. Note that ΔICP0 infection results in more abortive infections than does wildtype infection, manifesting in the presence of many ICP4+ cells that show diffused nuclear localizationand are unable to form replication compartments.
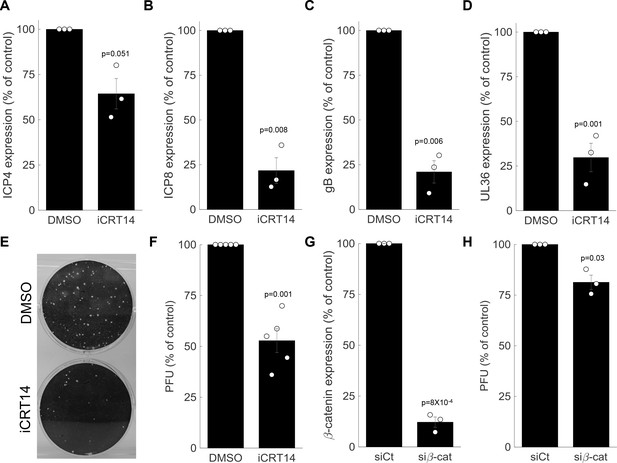
β-catenin is required for late viral gene expression and progeny production.
(A-D) HDFn treated with the β-catenin inhibitor iCRT14 or with vehicle alone (dimethyl sulfoxide (DMSO)) were infected with HSV-1 and analyzed for viral gene expression by RT-PCR at 5 hr post infection. Results show the means ± s.e. for three biological repeats, as compared to the DMSO treatment. The genes analyzed were ICP4 (A), ICP8 (B), gB (C) and UL36 (D). Circles are the results of individual experiments. p-values were calculated using a two-tailed one-sample t-test. (E) Representative image of a plaque assay, titrating virus generated from a 24-hr infection of HDFn treated with iCRT14 or DMSO. (F) Titers were calculated for viral progeny produced from iCRT14 or DMSO treated as in panel (E). The bars show the means ±s.e. from five biological repeats, as compared to the DMSO treatment. Circles are the results of individual experiments. p-values were calculated using a two-tailed one-sample t-test. (G,H) HDFn were treated with control short interfering RNA (siRNA) (siCt) or siRNA targeting β-catenin (siβ-cat) for 72 hr. Cells were assessed for β-catenin expression (G) or were infected for 24 hr before viruses were harvested and titrated as in panel (E) (H). Bars show the means ±s.e. from three biological repeats. p-values were calculated using a two-tailed one-sample t-test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HDFn, primary human dermal fibrobalsts | Cascade Biologics | cat #C0045C | |
| Cell line (Homo sapiens) | A549 | Sigma-Aldrich | cat #86012804-1VL | |
| Cell line (Homo sapiens) | Mel624 | Mel624 is a patient-derived melanoma cell-line, generated by the lab of Professor Thomas Gajewski at the Univeristy of Chicago | ||
| Cell line (Cercopithecus aethiops) | Vero | Obtained from the lab of Matthew D Weitzman, University of Pennsylvania | Used to grow wildtype HSV-1 and for plaque assay | |
| Cell line (Homo sapiens) | U2OS | Obtained from the lab of Matthew D Weitzman, University of Pennsylvania | Used to grow δICP0 HSV-1 | |
| Strain, strain background (HSV-1) | Wild-type strain 17, ICP4-YFP | Everett et al., 2003 | Obtained from the lab of Matthew D Weitzman, University of Pennsylvania | |
| Strain, strain background (HSV-1) | δICP0 strain 17, ICP4-YFP | Everett et al., 2003 | Obtained from the lab of Matthew D Weitzman, University of Pennsylvania | |
| Antibody | β-catenin (mouse monoclonal) | R and D systems | MAB13291-SP | Used for IF 1:400 |
| Antibody | IRF3 (rabbit monoclonal) | Cell Signaling Technologies | cat #11904 | Used for IF 1:200 |
| Sequence-based reagent | QPCR primer ICP4 Fwd | IDT | GCGTCGTCGAGGTCGT | |
| Sequence-based reagent | QPCR primer ICP4 Rev | IDT | CGCGGAGACGGAGGAG | |
| Sequence-based reagent | QPCR primer ICP8 Fwd | IDT | CGACAGTAACGCCAGAAGCTC | |
| Sequence-based reagent | QPCR primer ICP8 Rev | IDT | GGAGACAAAGCCCAAGACGG | |
| Sequence-based reagent | QPCR primer gB Fwd | IDT | CACCGCTACTCCCAGTTTATGG | |
| Sequence-based reagent | QPCR primer gB Rev | IDT | CCCTTGGCGTTGATCTTGTC | |
| Sequence-based reagent | QPCR primer UL36 Fwd | IDT | CGGGTCAAAAAGGTATGCGGTGT | |
| Sequence-based reagent | QPCR primer UL36 Rev | IDT | TGTCGTACACGCTCCTAACCATTG | |
| Sequence-based reagent | QPCR primer IFIT1 Fwd | IDT | CCT CCT TGG GTT CGT CTA CA | |
| Sequence- based reagent | QPCR primer IFIT1 Rev | IDT | GAA ATG AAA TGT GAA AGT GGC TGA T | |
| Sequence- based reagent | QPCR primer IFIT2 Fwd | IDT | GCTGAATCCTGACAACCAGTACC | |
| Sequence- based reagent | QPCR primer IFIT2 Rev | IDT | CACCTTCCTCTTCACCTTCTTCAC | |
| Sequence-based reagent | QPCR primer CTNNB1 Fwd | IDT | GAGATGGCCCAGAATGCAGTT | |
| Sequence-based reagent | QPCR primer CTNNB1 Rev | IDT | GGTGCATGATTTGCGGGAC | |
| Sequence-based reagent | siRNA againstβ-catenin | Dharmacon | M-003482-00-0005 | |
| Sequence-based reagent | siRNA non-targeting | Dharmacon | D-001206-13-05 | |
| Commercial assay or kit | RNEasy PLUS minikit | QIAGEN | cat #74134 | |
| Chemical compound, drug | iCRT14 | Sigma-Aldrich | cat ##SML0203 | Stock made in DMSO - 20 mM. Used at 20 micromolar final concentration |
| Software, algorithm | Single cell RNA seq analysis | This paper | https://github.com/nirdrayman/single-cell-RNAseq-HSV1.git |
Additional files
-
Supplementary file 1
Differential gene expression identified by sRNAseq.
(A) Genes that are upregulated in highly infected cells (wildtype infection). (B) GO annotations associated with genes from tab (A). (C) Transcription factors enriched in the promoters of genes from tab (A). (D) Genes that are upregulated in highly infected cells (ΔICP0 infection). (E) GO annotations associated with genes from tab (D). (F) Transcription factors enriched in the promoters of genes from tab (D).
- https://doi.org/10.7554/eLife.46339.019
-
Supplementary file 2
Analysis of genes that are upregulated in ICP4-negative sorted cells.
(A) Genes that are upregulated in ICP4-negative cells (afer wildtype infection). (B) GO annotations associated with genes from tab (A). (C) Genes that are upregulated in ICP4-negative cells (after ΔICP0 infection). (D) GO annotations associated with genes from tab (C). (E) Transcription factors enriched in the promoters of genes from tab (C).
- https://doi.org/10.7554/eLife.46339.020
-
Supplementary file 3
Analysis of genes upregulated in ICP4-positive sorted cells.
(A) Genes that are upregulated in ICP4-positive cells (after wildtype infection). (B) GO annotations associated with genes from tab (A). (C) Transcription factors that are enriched in the promoters of genes from tab (A). (D) Genes that are upregulated in ICP4-positive cells (after ΔICP0 infection). (E) GO annotations associated with genes from tab (D). (F) Transcription factors that are enriched in the promoters of genes from tab (D).
- https://doi.org/10.7554/eLife.46339.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46339.022





