Metabolic constraints drive self-organization of specialized cell groups
Figures
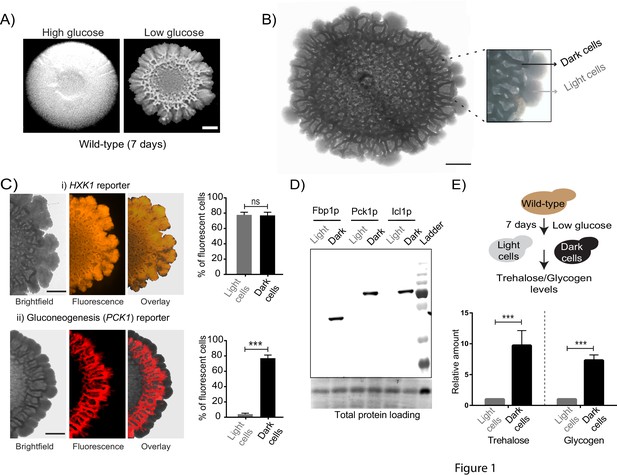
Cells within S.cerevisiae colonies exhibit ordered metabolic specialization.
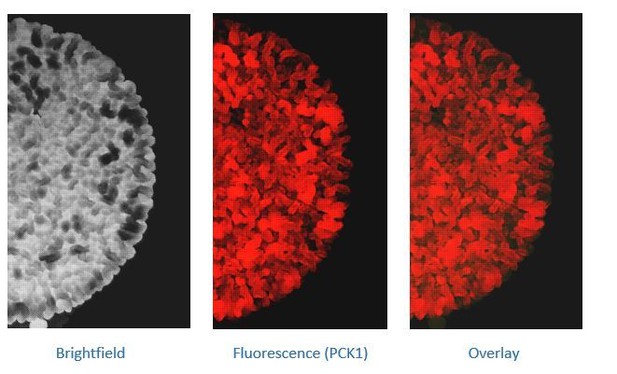
(A) Low glucose is required for rugose colonies to develop. The panel shows the morphology of mature yeast colonies in rich medium, with supplemented glucose as the sole variable. Scale bar: 2 mm. (B) Reconstructed bright-field images of a mature wild-type colony. Within the colony, a network of dark and bright regions is clearly visible, as classified based purely on optical density. We classify the cells in the dark region as dark cells, and in the peripheral light region as light cells. Scale bar: 2 mm. (C) Spatial distribution of mCherry fluorescence across a colony, indicating the activity of (i) a reporter for hexokinase (HXK1) activity, or (ii) a gluconeogenesis dependent reporter (PCK1), in two different colonies. The percentage of fluorescent cells (in isolated light and dark cells from the respective colonies) were also estimated by flow cytometry, and is shown as bar graphs. Scale bar: 2 mm. Also see Figure 1—figure supplement 1A–B and Figure 1—figure supplement 2A for more information. (D) Western blot based detection of proteins involved in gluconeogenesis (Fbp1p and Pck1p), or associated with increased gluconeogenic activity (Icl1p), in isolated dark or light cells. The blot is representative of three biological replicates (n = 3). Also see Figure 1—figure supplement 2B for more information. (E) Comparative steady-state amounts of trehalose and glycogen (as gluconeogenesis end point metabolites), in light and dark cells (n = 3). Statistical significance was calculated using unpaired t test (*** indicates p<0.001) and error bars represent standard deviation.
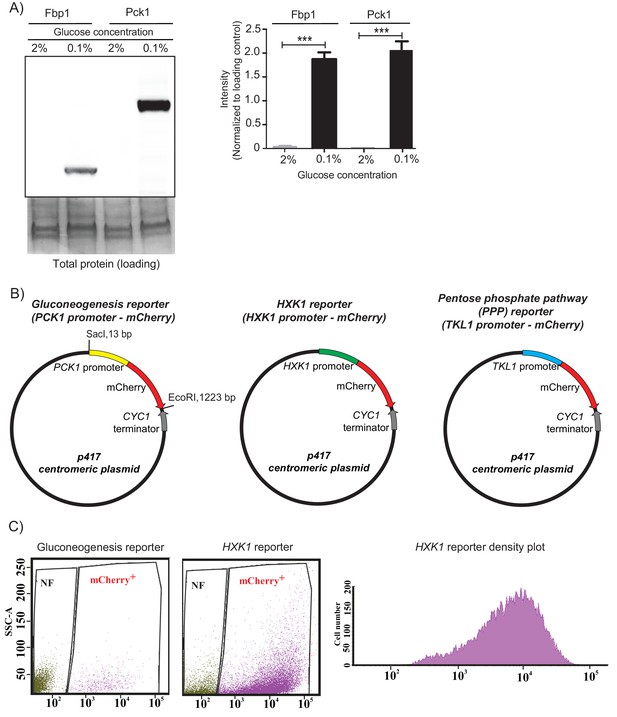
Gluconeogenesis activity is spatially restricted.
(A) Western blot based detection of proteins involved in gluconeogenesis (Fbp1p and Pck1p), from cells grown in high (2%) and low (0.1%) conditions. The blot is representative of three biological replicates (n = 3). Quantification of the band intensities were done using ImageJ and the intensity values were normalized to the protein loading controls. (B) Plasmid maps of the different reporters used in this study. Gene cassette containing the specific promoter and mCherry was cloned into the p417 centromeric plasmid using the SacI and EcoRI restriction sites. (C) Scatter plot depicting distribution of mCherry positive light cells isolated from wild-type colonies with the gluconeogenesis or HXK1 reporter. Density plot of mCherry positive light cells in the HXK1 reporter is shown. SSC-A represents side scatter measurement and NF represents non-fluorescent population.
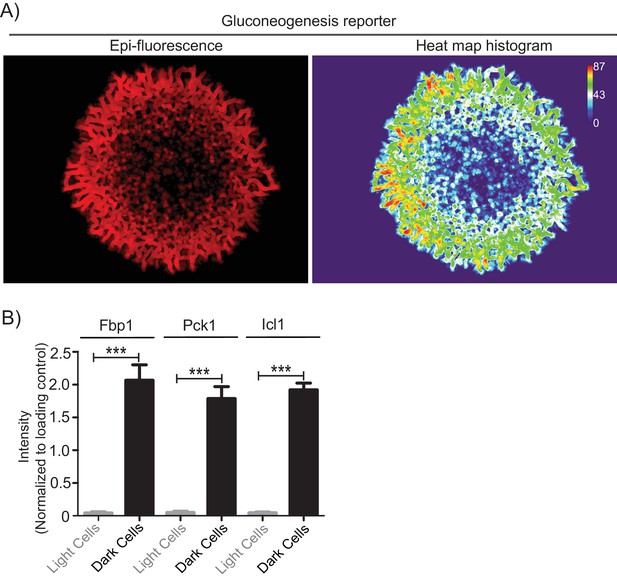
Gluconeogenesis activity is spatially restricted.
(A) Whole colony fluorescence image of wildtype colony with the gluconeogenesis reporter. Heat map histogram analysis of the fluorescent intensity of the same colony was done using ImageJ. Values represent pixel intensities.B) Quantification of the Fbp1, Pck1 and Icl1 protein levels from light and dark cells (Figure 1D). Quantification of the western blot band intensities were done using ImageJ and the intensity values were normalized to the protein loading controls. Statistical significance was calculated using unpaired t test (*** indicates p<0.001) and error bars represent standard deviation.
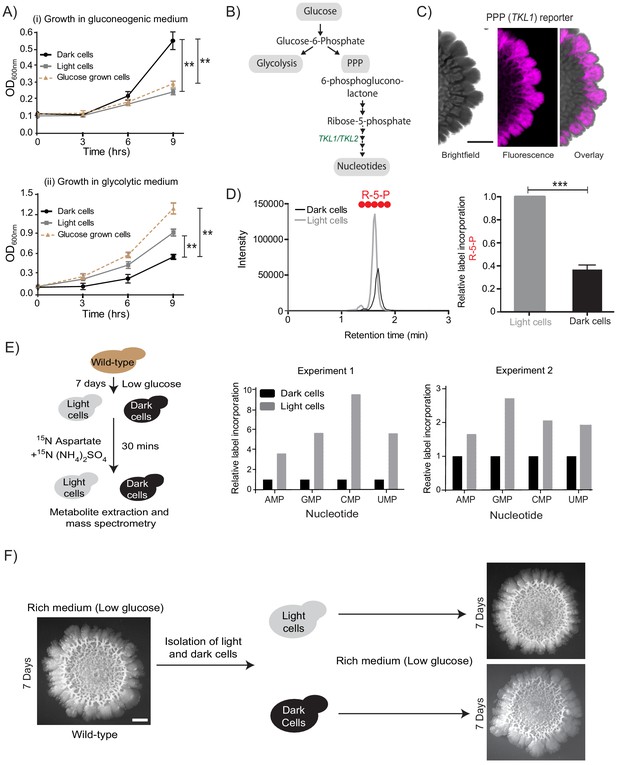
Cells organize into spatially restricted, contrary metabolic states within the colony.
(A) Comparative immediate growth of isolated light cells and dark cells, transferred to a ‘gluconeogenic medium’ (2% ethanol as carbon source), or a ‘glycolytic medium’ (2% glucose as carbon source), based on increased absorbance (OD600) in culture. Wild-type cells growing in liquid medium (2% glucose) in log phase (i.e. in a glycolytic state) were used as controls for growth comparison (n = 3). (B) A schematic showing metabolic flow in glycolysis and the pentose phosphate pathway (PPP), and also illustrating the synthesis of nucleotides (dependent upon pentose phosphate pathway). TKL1 controls an important step in the PPP, and is strongly induced during high PPP flux. (C) Spatial distribution of mCherry fluorescence across a colony, based on the activity of a PPP- dependent reporter. Scale bar: 2 mm. Also see Figure 1—figure supplement 1A and Figure 2—figure supplement 1A. (D) LC-MS/MS based metabolite analysis, using exogenously added 13C Glucose, to compare flux of 13C Glucose into the PPP metabolite ribulose-5-phosphate (R-5-P), in light and dark cells. The red circles represent 13C labeled carbon atoms (n = 3). Also see Figure 2—figure supplement 1B. (E) Comparative metabolic-flux based analysis comparing 15N incorporation into newly synthesized nucleotides, in dark and light cells. Also see S2, and Materials and methods. (F) Light cells and dark cells isolated from a 7 day old wild-type complex colony re-form indistinguishable mature colonies when re-seeded onto fresh agar plates, and allowed to develop for 7 days. Scale bar = 2 mm. Statistical significance was calculated using unpaired t test (*** indicates p<0.001, ** indicates p<0.01) and error bars represent standard deviation.
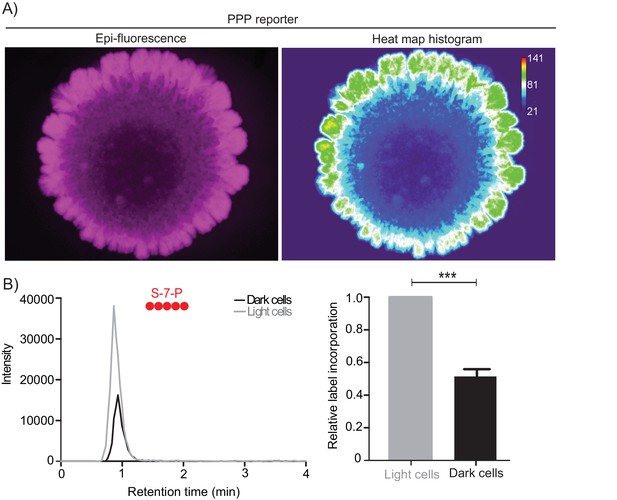
Light cells exhibit high PPP activity.
(A) Whole colony fluorescence image of wildtype colony with the pentose phosphate pathway (PPP) reporter. Heat map histogram analysis of the fluorescent intensity of the same colony was done using ImageJ. Values represent pixel intensities. (B) LC-MS/MS based metabolite analysis, using exogenously added 13C glucose, to compare flux of 13C glucose into the PPP metabolite sedoheptulose-7-phosphate (S-7-P), in light and dark cells. The red circles represent 13C labeled carbon atoms. Relative label incorporation of 13C labeled carbon atoms in dark cells with respect to light cells were quantified. Statistical significance was calculated using unpaired t tests (*** indicates p<0.001) and error bars represent standard deviation.
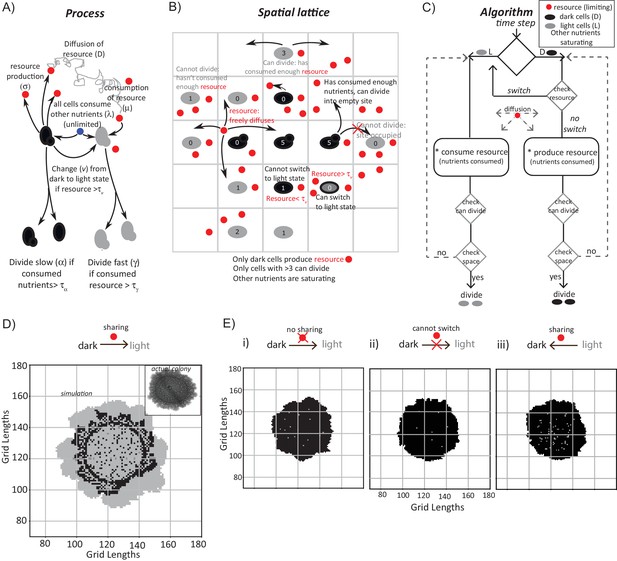
A mathematical model suggests constraints for the emergence and organization of cells in complimentary metabolic states.
(A) Processes, based on experimental data, incorporated into developing a simple mathematical model to simulate colony development. The dark and light cells are appropriately colored, and the parameters incorporated are resource production (σ), diffusion parameters for the resource (D), consumption of the resource (µ), and fast or slow rates of division (α or γ), based on resource or amino acid consumption. (B) The spatial distribution of cells is reduced to a grid like lattice within the model, to allow coarse graining of the location of cells across a colony. The rules for cell division and expansion incorporate the ability to consume existing nutrients in the medium, produce a resource and/or consume a produced resource, and a threshold amount of resource build up before utilization. (C) A flow-chart of the algorithm used in the mathematical model. The decision making process in the algorithm, incorporating all the elements described in panels (A) and (B) is illustrated. Also see Figure 3—figure supplement 1A–C and Materials and methods. (D) A simulation of the development of a wild-type colony, based on the default model developed. The inset shows an image of a real wild-type colony (same brightfield image used in Figure 1B), which has developed for an equivalent time (~6 days). Also see Figure 3—figure supplement 1A–C and Video 1. (E) A simulation of colony development using the model, where key parameters have been altered. (i) The sharing of a produced resource is restricted. (ii) The ability to switch from a dark to a light state is restricted. (iii) Light cells produce a resource taken up by dark cells is included. Note that in all three scenarios the colony size remains small, and fairly static. Also see Figure 3—figure supplement 2A–D and Videos 2, 3 and 4.
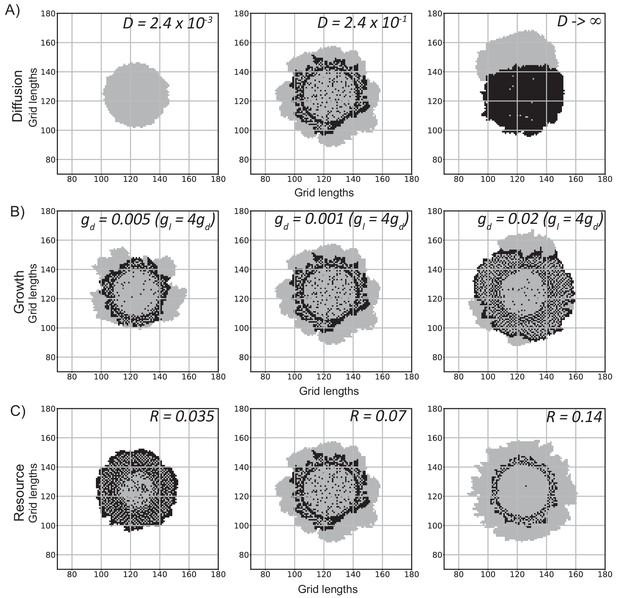
Effects on the colony as we change individual parameters used in the model.
(A) Changing the diffusion constant (D [L2T−1]) of the resource in the medium. The diffusion constant used to simulate the left panel colony is 100 times smaller than the default colony (middle panel). The right panel colony was simulated with a very large effective diffusion constant. To get this fast diffusion, the total available resource was uniformly redistributed over the grid between every two time steps. Thus, every grid location had the same level of resource before the cells consume or produce it, as if the resource diffused very quickly over the grid. (B) Changing the growth rate of the cells. gd and gl are growth rate parameters of the dark and light cells respectively. The light cells grow four times faster than the dark cells in all cases. The middle panel colony is the default colony. gd = 0.01. The one on the left has half the growth rate and the one on the right has twice the growth rate in comparison. (C) Changing the resource (R) produced by the dark cells. The light cells consume resource at a fixed rate if available. The middle panel has R = 0.07. The left colony simulation has half the produced value and the right colony simulation has double the produced value. All colony simulations were started with 1257 grids (each containing ~100 cells), of which 99% were dark cells, and were run for 750 time steps.
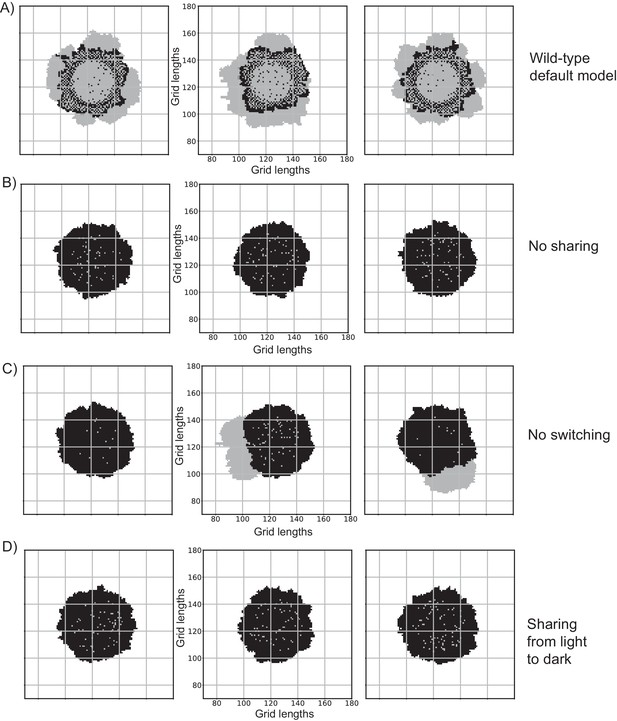
Reproducibility of the model, under different scenarios.
Since the colony simulations have elements of stochasticity present in the model, presented are a few replicate colonies from independent simulation. These showcase crucial and reproducible aspects of the different conditions explored in Figure 3D and Figure 3E. (A) three wild-type colonies generated with the default parameter set. The common features are a circular centre with mostly light cells, an annular region with dark cells and a few light cells, and the colony periphery with mostly light cells. (B) three replicate colonies with the ‘no sharing’ condition. The dark cells do not produce any shared resource. The final colonies have mostly dark cells as the number of light cells doesn't change. (C) three replicate colonies with the ‘no switching’ condition. If there are a few light cells at the periphery, they can consume the shared resource produced by the dark cells and grow into the empty space. However, the centre of the colonies has mostly dark cells. (D) three replicate colonies with the ‘wrong sharing’ condition (sharing of a resource made by light cells, taken up by dark cells), which are similar to the ‘no sharing’ condition.
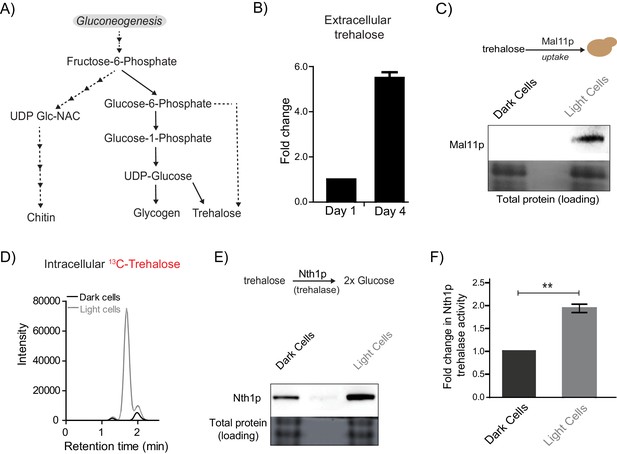
Trehalose satisfies criteria to be the metabolic resource determining the emergence of light cells.
(A) A schematic illustrating the metabolic intermediates and different end-point metabolites of gluconeogenesis. (B) Extracellular amounts of trehalose measured from developing wild-type colonies. Entire colonies were isolated, and only exogenous trehalose estimated, at the respective days. Fold change in the amount of extracellular trehalose produced by a 4 day old colony with respect to a 1 day old colony was calculated (n = 3). (C) Comparative protein amounts of Mal11, a major transporter of trehalose in S. cerevisiae, in light and dark cells, as measured using a Western blot is shown. The blot is representative of 3 independent experiments (n = 3). (D) Estimates of the relative ability of light and dark cells to uptake trehalose is shown. 13C Trehalose was exogenously added to light and dark cells, and intracellular amounts of the same are shown (as intensity of the MS/MS peak corresponding to 13C-trehalose) (n = 3). (E) Comparative amounts of Nth1, the major intracellular trehalase enzyme in S. cerevisiae, in light and dark cells, as measured using a Western blot is shown. The blot is representative of 3 independent experiments (n = 3). (F) in vitro neutral trehalase activity present in lysed light or dark cells is shown (n = 3). Statistical significance was calculated using unpaired t test (** indicates p<0.01) and error bars represent standard deviation.
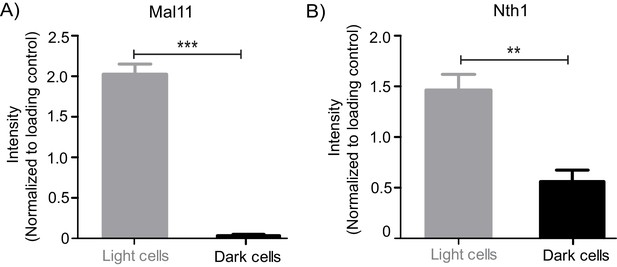
Quantitation of relative Mal11 and Nth1 protein levels in light and dark cells.
(A) Quantification of the Mal11 protein levels from light and dark cells (Figure 4C). Quantification of the western blot band intensities were done using ImageJ and the intensity values were normalized to the protein loading controls. (B) Quantification of the Nth1 protein levels from light and dark cells (Figure 4E). Quantification of the western blot band intensities were done using ImageJ and the intensity values were normalized to the protein loading controls. Statistical significance was calculated using unpaired t test (*** indicates p<0.001, ** indicates p<0.01) and error bars represent standard deviation.
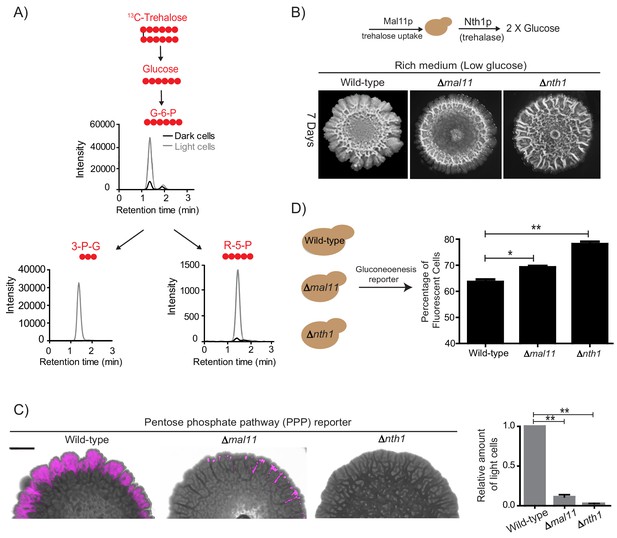
Trehalose uptake and utilization determines the existence of light cells.
(A) Estimation of trehalose uptake and breakdown/utilization in light and dark cells. LC-MS/MS based metabolite analysis, using exogenously added 13C Trehalose, to compare breakdown and utilization of 13C Trehalose for glycolysis and the PPP, in light and dark cells. The red circles represent 13C labeled carbon atoms. Data for 13C labeled glycolytic and PPP intermediates (derived from trehalose) are shown. The data presented is from a single flux experiment, which was repeated independently (with different colonies) twice (n = 2). Also see Figure 5—figure supplement 1A. (B) Comparative development of wild-type colonies (same image used in Figure 1A) with colonies lacking the major trehalose transporter (∆mal11), or the intracellular neutral trehalase (∆nth1). Colonies are shown after 7 days of development. Scale bar: 2 mm. (C) Visualization (left panel) and quantification (right bar graphs) of light cells in wild-type (same image used in Figure 2C), ∆mal11, or ∆nth1 cells, based on fluorescence emission dependent upon the PPP reporter activity. The quantification is based on flow cytometry data (n = 3). Scale bar: 2 mm. (D) Estimate of the percentage of gluconeogenic cells in wild-type, ∆mal11 and ∆nth1 (strains that cannot up-take or breakdown trehalose). This was based on quantifying the expression of the gluconeogenesis reporter plasmid (pPCK1-mCherry), expressed in all these cells. Cells from the entire colony were isolated and percentage of fluorescent cells (i.e. cells expressing the gluconeogenic reporter) in each colony was calculated by analyzing the samples by flow cytometry (n = 3). Statistical significance was calculated using unpaired t test (* indicates p<0.05, ** indicates p<0.01) and error bars represent standard deviation.
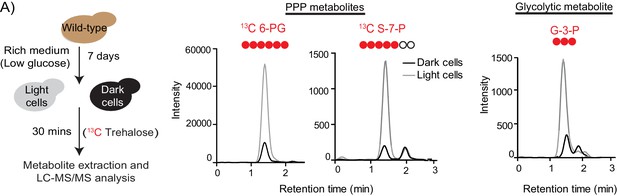
Comparative breakdown of labeled trehalose by distinct cells in a colony.
(A) Trehalose breakdown by light cells and dark cells were monitored by isolating light and dark cells from a ~ 5 to 6 day old wild-type colony and incubating them briefly (30 min) with labeled 13C Trehalose. Post incubation, metabolites from light and dark cells were extracted and analyzed for the pentose phosphate pathway (PPP) intermediates 13C 6-phosphogluconate (13C 6 PG), 13C Sedoheptulose-7-phosphate (13C S-7-P) and glycolytic intermediate 13C Glyceraldehyde-3-phosphate (13C G-3-P) using LC-MS/MS.
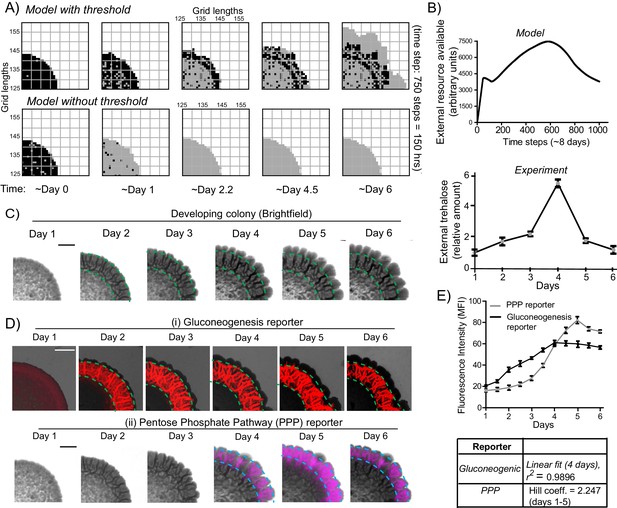
A resource threshold effect controls cooperative switching of cells to the light state.
(A) Simulation of colony development, based on the default model (which incorporates a resource threshold buildup, followed by consumption, switching to a light state, and expansion), compared to a model where the threshold amounts of the resource is removed. Note the final expansion size of the colony. Also see Figure 6—figure supplement 1A–D and Video 5. (B) (i) Changes in the availability of the resource as the colony develops, based on the model. (Ii) Extracellular amounts of trehalose measured from developing wild-type colonies. Data from three independent colonies. Note: in the model, in ~3–4 days the resource is highest, and reduces sharply after that. In the experimentally obtained data, extracellular trehalose amounts are highest at ~day 4, and then rapidly decreases over day 5. This correlates to when the light cells emerge and expand. (C) A time-course of bright-field images of the developing wild-type colony, illustrating the distribution of dark cells, and the emergence and distribution of light cells. (D) A time-course revealing fluorescence based estimation of the (i) reporter for gluconeogenic activity (dark cells), or (ii) the PPP activity reporter (light cells). Note the delayed, rapid appearance and increase in the PPP activity reporter. (E) Quantification of the increase in the gluconeogenic reporter activity in the colony, and the PPP reporter activity (based on fluorescence intensity) within the colony. The increase in gluconeogenic reporter activity, when plotted, is linear, and saturates. The increase in PPP activity over the first 5 days is highly cooperative (as estimated using a Hill coefficient as a proxy for cooperativity), before saturating (n = 3). Error bars represent standard deviation.
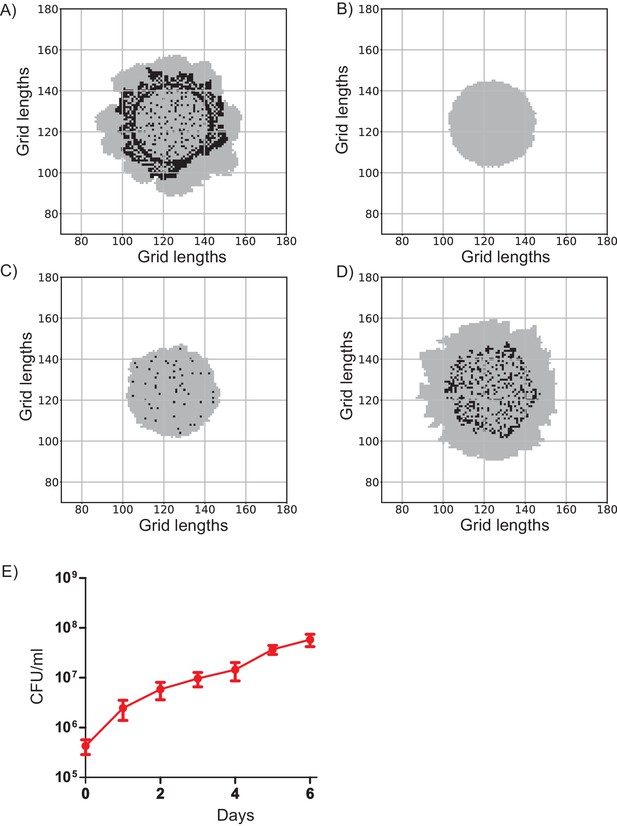
Colonies generated using different switching rules.
(A) Wild-type colony with a resource threshold switching rule. In every time step, the algorithm checks if the shared resource levels at the locations of dark cells are more than a threshold value of 3 units. If so, there is a 50% chance of those cells switching to light cells. (B) A colony generated without the need for a resource threshold level. Dark cells switch by random chance. Light cells cannot switch to dark. (C and D) Colonies generated using a linear switching rule. The probability of switching increases linearly with the locally available resource levels till a maximum value of 1.0. The probability of switching is of the form p=mU (if mU less than 1.0) and p=1.0 otherwise. Here U is the resource level at the location of the dark cells and m is the slope of the linear relationship. In panel (C) m = 1. In panel (D) m = 0.01. (E) Total number of viable cells in a developing colony was estimated over time. CFU denotes colony forming units. Error bars represent standard deviation. Video clip legends.
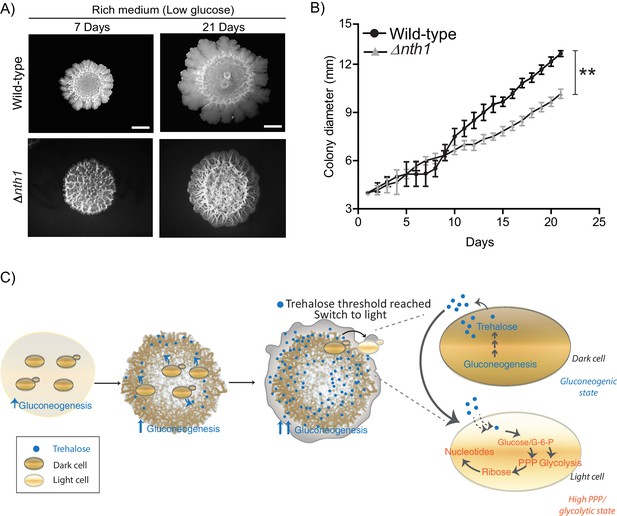
Physiological advantages of cells with organized spatial heterogeneity.
(A) Foraging response of wild-type cells (same image used in Figure 1A) and Δnth1 cells measured as a function of their ability to spread on a plate. Colony spreading was quantified by measuring the diameter of the colonies every day for 21 days (n = 3). (B) Cells in low glucose perform gluconeogenesis, as required in low glucose medium. As gluconeogenic reserves build up, trehalose builds up in the extracellular environment. At a threshold concentration of trehalose, some cells switch to a high glycolytic, PPP state. This state depends upon the utilization of trehalose to fuel it. This utilization of trehalose by the light cells results in decreased external trehalose to below a threshold. This in turn restrains the other, remaining cells in a gluconeogenic state, where they continue to produce trehalose. This gives rise to the final, self-organized community, with specialization of function and division of labor. Statistical significance was calculated using unpaired t test (** indicates p<0.01) and error bars represent standard deviation.
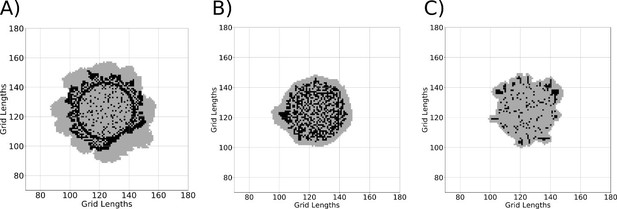
A simulation of colony development using the model using different concentrations of gluconeogenic cells A) 95% B) 50% C) 10% during the start of colony development.
Black regions represent dark cells and grey regions represent light cells.
Videos
Development of the colony.
Simulation video showing the changes in a wild-type model colony. After a small lag, dark cells at the edge start dividing into empty space and due to the threshold switching effect, the center of the colony now has light cells. Cells at the periphery also switch to light cells which divide faster by utilizing resource shared by the dark cells.
Dark cells do not share the metabolic resource with light cells.
Simulation video showing changes in a colony where the dark cells don't share any resource for the light cells to consume. The number of light cells doesn't increase and the final colonies predominantly comprise of dark cells.
Dark cells do not switch to light cells.
Simulation video showing changes in a colony where the dark cells don't switch to light cells but continue to produce resource on to the resource grid. In certain cases, there can be light cells at the very edge of the starting colony. This is because the composition of the colonies might be the same between simulations, but individual cell block locations are done at random. Such cells at the edge can utilize the shared resource and divide into empty space on the grid.
Light cells share metabolic resource with dark cells (Wrong sharing).
Simulation video showing changes in a colony where the dark cells don't share any resource for the light cells but the light cells provide amino acids for the dark cells to consume (wrong sharing). The dark cells have an abundance of amino acids to grow and divide. The final colonies predominantly comprise of dark cells.
Development of colony without a resource threshold.
Simulation video showing changes in a colony where the dark cells switch to light by random chance (probability p=0.5). They don't need the resource levels to reach a certain threshold. Once they become light cells, they cannot switch back to dark. Due to lack of the produced resource, the colony doesn't grow discernibly.
Tables
Mass transitions used for LC-MS/MS experiments.
| Nucleotides | Formula | Parent/Product (positive polarity) | Comment (for 15N experiment) |
|---|---|---|---|
| AMP | C10H14N5O7P | 348/136 | Product has all N |
| 15N_AMP_1 | 349/137 | ||
| 15N_AMP_2 | 350/138 | ||
| 15N_AMP_3 | 351/139 | ||
| 15N_AMP_4 | 352/140 | ||
| 15N_AMP_5 | 353/141 | ||
| GMP | C10H14N5O8P | 364/152 | Product has all N |
| 15N_GMP_1 | 365/153 | ||
| 15N_GMP_2 | 366/154 | ||
| 15N_GMP_3 | 367/155 | ||
| 15N_GMP_4 | 368/156 | ||
| 15N_GMP_5 | 369/157 | ||
| CMP | C9H14N3O8P | 324/112 | Product has all N |
| 15N_CMP_1 | 325/113 | ||
| 15N_CMP_2 | 326/114 | ||
| 15N_CMP_3 | 327/115 | ||
| UMP | C9H13N2O9P | 325/113 | Product has all N |
| 15N_UMP_1 | 326/114 | ||
| 15N_UMP_2 | 327/115 | ||
| Trehalose and sugar phosphates | Formula | Parent/Product (negative polarity) | Comment (for 13C experiment) |
| Trehalose | C12H22O11 | 341.3/179.3 | |
| 13C_Trehalose_12 | 353.3/185.3 | Product has 6 C all of which are labeled | |
| G3P | C3H7O6P | 169/97 | Monitoring the phosphate release |
| 13C_G3P_3 | 172/97 | ||
| 3 PG | C3H7O7P | 185/97 | Monitoring the phosphate release |
| 13C_3 PG_3 | 188/97 | ||
| G6P | C6H13O9P | 259/97 | Monitoring the phosphate release |
| 13C_G6P_6 | 265/97 | ||
| 6 PG | C6H13O10P | 275/97 | Monitoring the phosphate release |
| 13C_6 PG_6 | 281/97 | ||
| R5P | C5H11O8P | 229/97 | Monitoring the phosphate release |
| 13C_R5P_5 | 234/97 | ||
| S7P | C7H15O10P | 289/97 | Monitoring the phosphate release |
| 13C_S7P_5 | 294/97 |
Strains and plasmids used in this study.
| Strain/genotype | Information | Source/reference |
|---|---|---|
| Wild-type (WT) | YBC16G1, prototrophic sigma1278b, MAT a | Isolate via Fink Lab |
| WT (pPCK1-mCherry) | Wild-type strain with gluconeogenesis reporter plasmid (mCherry with PCK1 promoter) | this study |
| WT (pHXK1-mCherry) | Wild-type strain with constitutive reporter plasmid (mCherry with HXK1 promoter) | this study |
| WT (pTKL1-mCherry) | Wild-type strain with pentose phosphate pathway reporter plasmid (mCherry with TKL1 promoter) | this study |
| PCK1-flag | MAT a PCK1-3xFLAG::natNT2 | this study |
| FBP1-flag | MAT a FBP1-3xFLAG::natNT2 | this study |
| ICL1-flag | MAT a ICL1-3xFLAG::natNT2 | this study |
| MAL11-flag | MAT a MAL11-3xFLAG::natNT2 | this study |
| NTH1-flag | MAT a NTH1-3xFLAG::natNT2 | this study |
| ∆nth1 | MAT a nth1::kanMX6 | this study |
| ∆mal11 | MAT a mal11::kanMX6 | this study |
| ∆nth1 (pPCK1-mCherry) | ∆nth1 strain with gluconeogenesis reporter plasmid (mCherry with PCK1 promoter) | this study |
| ∆mal11 (pPCK1-mCherry) | ∆mal11 strain with gluconeogenesis reporter plasmid (mCherry with PCK1 promoter) | this study |
| ∆nth1 (pTKL1-mCherry) | ∆nth1 strain with pentose phosphate pathway reporter plasmid (mCherry with TKL1 promoter) | this study |
| ∆mal11 (pTKL1-mCherry) | ∆mal11 strain with pentose phosphate pathway reporter plasmid (mCherry with TKL1 promoter) | this study |
| Plasmid | Information | Source/reference |
| pPCK1-mCherry | mCherry under the PCK1 promoter and CYC1 termin- ator. p417 centromeric plasmid backbone, G418R. | this study |
| pHXK1-mCherry | mCherry under the HXK1 promoter and CYC1 termin- ator. p417 centromeric plasmid backbone, G418R. | this study |
| pTKL1-mCherry | mCherry under the TKL1 promoter and CYC1 termin- ator. p417 centromeric plasmid backbone, G418R. | this study |
Parameters of the model for the wild-type case.
| Parameter | Notation | Value |
|---|---|---|
| Growth rate (dark cell block) | gd | 0.01/T |
| Growth rate (light cell block) | gl | 0.04/T |
| Switching threshold | S | 3.0 units |
| Resource produced by each dark cell block | R | 0.07 units/T |
| Resource or amino acids consumed per cell block (light or dark) | C | 0.05 units/T |
| Minimum resource or amino acid reserve needed for division (light or dark) | -- | 1.0 unit |
| Chance to switch to light cell if threshold reached | P | 0.5/T |
| Diffusion constant of the resource | D | 0.24 L2/T |
Additional files
-
Source code 1
Simulation codes in Python.
- https://cdn.elifesciences.org/articles/46735/elife-46735-code1-v3.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/46735/elife-46735-transrepform-v3.docx