Structure-based inhibitors of amyloid beta core suggest a common interface with tau
Figures
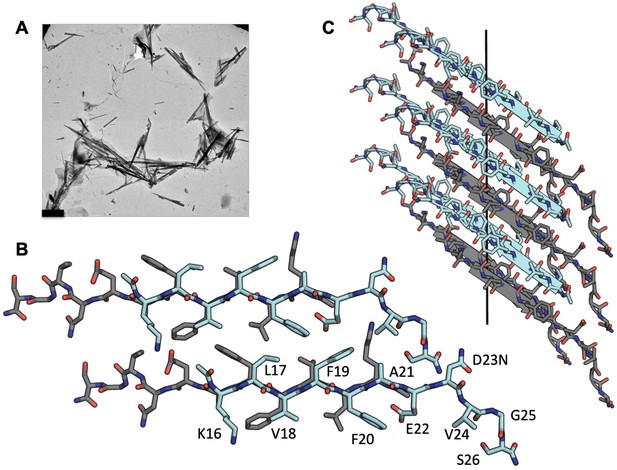
MicroED structure of segment Aβ 16–26 D23N from microcrystals.
(A) Electron micrograph of 3D crystals used for data collection, scale bar is 1 µm. (B) The crystal structure reveals tightly mated pairs of anti-parallel β-sheets with opposing sheets in gray and cyan. The side-chains interdigitate to form a dry interface. Two neighboring sheets are viewed perpendicular to the β-sheets. (C) View of 6 layers perpendicular to the fibril axis (black line). The β-sheets stack out of register along the fibril axis.
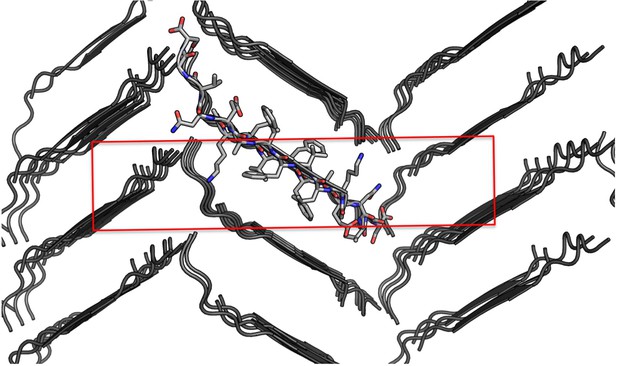
Crystal packing of the Aβ 16–26 D23N atomic structure.
View down the ‘a’ axis of the unit cell, outlined in red.
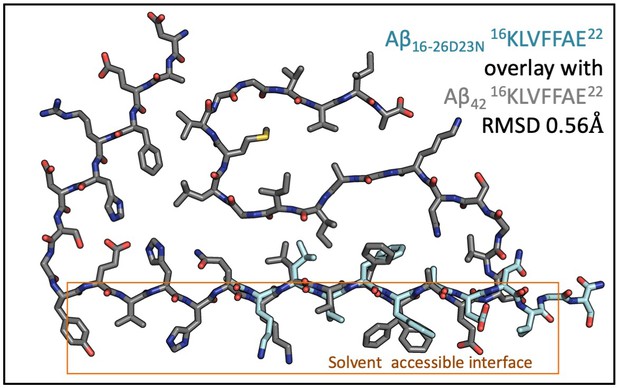
The spines of Aβ 16–26 D23N and Aβ1-42 fibrils (5OQV) are structurally similar.
32 backbone atoms differ from each other by 0.57 Å RMSD. RMSD values were calculated using LSQ in Coot.
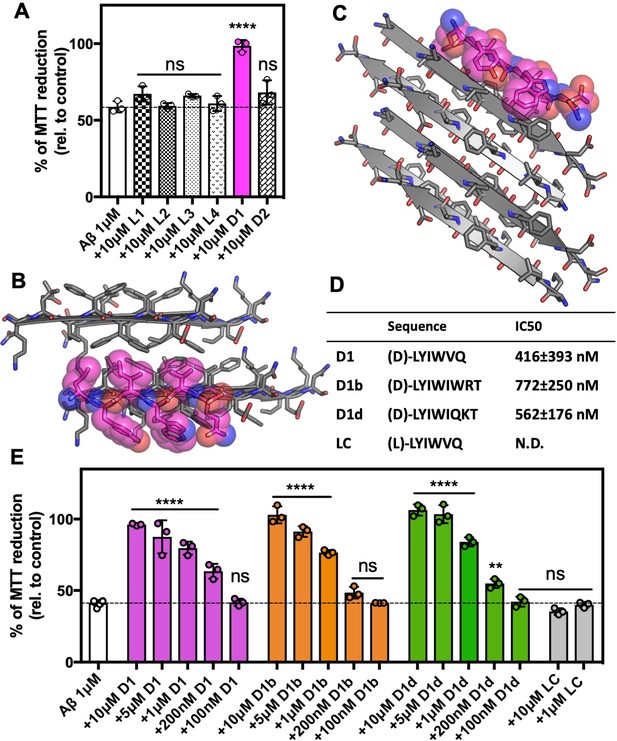
Development of inhibitors of Aβ fibril formation using structure-based design against Aβ 16–26 D23N.
(A) Identification of Aβ1–42 inhibitor. 10 μM Aβ1–42 was incubated alone or with 100 μM of each candidate peptide inhibitor for 12 hr at 37°C and then diluted 1:10 with pre-plated N2a cells. Cytotoxicity was quantified using MTT dye reduction Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3; ns = not significant; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column) (B, C) Segment KLVFFAEN, derived from the Aβ 16–26 D23N crystal structure, was used as the design target. Model of peptide inhibitor D1(magenta) bound to the design target, KLVFFAEN (gray). Smaller hydrophobic residues of D1 mimic interactions with the fibril interface on one side of the peptide (B), whereas the other side of the peptide positions large aromatic residues between Aβ residues, breaking possible further interactions (C). (D) Overview of peptide inhibitors in D and L amino acid conformations, as indicated, used in this study and their sequences. Peptide LC is the L-form cognate peptide of D-form peptide D1 and is the negative control for peptide inhibitor D1 and its derivatives D1b and D1d. IC50 values were determined using four parameter nonlinear fit for half maximal inhibition. N.D., not determined. (E) Peptide inhibitors D1, D1b, and D1d reduce the cytotoxicity of Aβ1–42 in a dose dependent manner, whereas control peptide LC does not. 10 μM Aβ1–42 was incubated alone or with various concentrations of each peptide inhibitor for 12 hr at 37°C and then diluted 1:10 with pre-plated N2a cells. Cytotoxicity was quantified using MTT dye reduction. Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3–6; ns = not significant; **, p<0.002; ****, p<0.0001 using an ordinary one-way ANOVA Dunnett’s relative to leftmost column).
-
Figure 2—source data 1
Toxicity data points—Aβ incubated with inhibitors and inhibitor controls.
- https://doi.org/10.7554/eLife.46924.008
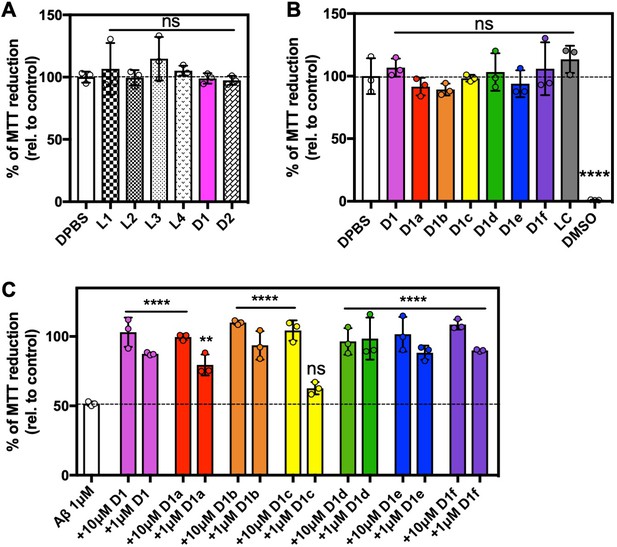
Extended Toxicity data.
(A, B) Peptide inhibitors are not toxic. 100 μM of each peptide inhibitor was incubated for 12 hr at 37°C and then diluted 1:10 with pre-plated N2a cells. Cytotoxicity was quantified using MTT dye reduction. Bars represent mean with individual technical replicates, error bars display one standard deviation. (C) Second generation peptide inhibitors reduce the cytotoxicity of Aβ1–42. 10 μM Aβ1–42 was incubated alone or with 10 μM and 100 μM of each peptide inhibitor for 12 hr at 37°C and then diluted 1:10 with pre-plated N2a cells. Cytotoxicity was quantified using MTT dye reduction. Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3; ns = not significant; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column).
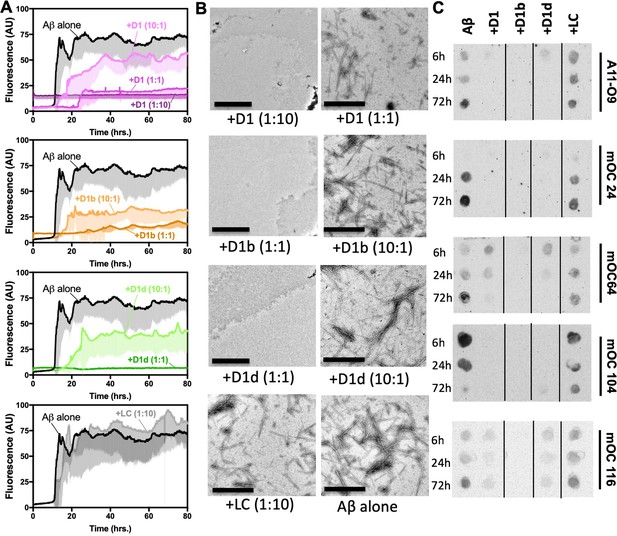
Designed inhibitors reduce aggregation of Aβ1–42.
(A) Peptide inhibitors D1, D1b, and D1d reduce fibril formation of Aβ1–42, while negative control peptide LC does not. 10 μM of Aβ1–42 was incubated alone or at a 1:10, 1:1, or 10:1 molar ratio to each inhibitor under quiescent conditions at 37°C. Fibril formation was monitored using ThT fluorescence. Curves show the average of three technical replicates with one standard deviation below. (B) Negative-stain TEM analysis confirms the results of the ThT assays in Figure 3A. Samples were prepared as above and incubated for 72 hr before TEM analysis. Images of Aβ1–42 to D1 (1:10), D1b (1:1) and D1d (1:1) were captured at 3200x; scale bars are 2 µm. All other images were captured at 24,000x; scale bars are 500 nm. (C) Peptide inhibitors reduce the formation of Aβ1–42 assemblies recognized by conformational monoclonal antibodies, while negative control peptides do not. 10 μM Aβ1–42 was incubated alone (left-most column) or with 10-fold molar excess of each peptide-based inhibitor. Aliquots of the reaction were tested for antibody-binding at 6 hr, 24 hr, and 72 hr. Membranes were spliced as indicated for clarity.
-
Figure 3—source data 1
Dot blot quatification data points.
- https://doi.org/10.7554/eLife.46924.012
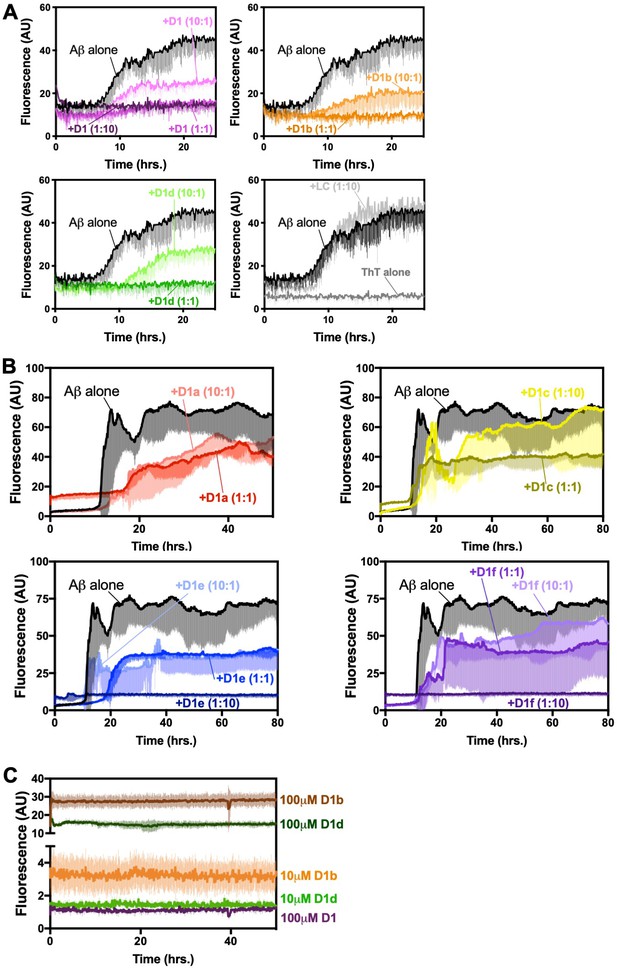
A Peptide inhibitors D1, D1b, and D1d reduce fibril formation of Aβ1–42, while negative control peptide LC does not.
10 μM of Aβ1–42 was incubated alone or at a 1:10, 1:1, or 10:1 molar ratio to each inhibitor under quiescent conditions at 37°C in polypropylene plates. Fibril formation was monitored using ThT fluorescence. Curves show the average of three technical replicates with one standard deviation below. (B) Peptide inhibitors D1a, D1c, D1e, and D1f are less effective ate reducing fibril formation of Aβ1–42, 10 μM of Aβ1–42 was incubated alone or at a 1:10, 1:1, or 10:1 molar ratio to each inhibitor under quiescent conditions at 37°C. Fibril formation was monitored using ThT fluorescence. Lines show the average of three technical replicates with one standard deviation below. Inhibitors D1a and D1c are not shown at (1:10) ratio due to high ThT signal from peptide self-association. (C) Peptide inhibitors D1b and D1d self-associate at high concentrations. Peptide inhibitors were incubated at 10 μM and 100 μM under quiescent conditions at 37°C. Fibril formation was monitored using ThT fluorescence. Lines show the average of three technical replicates with one standard deviation displayed below.
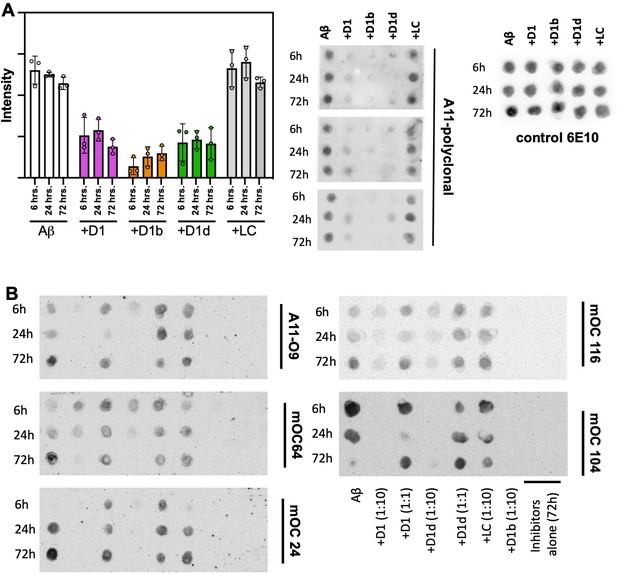
Extended dot blot data.
(A) Peptide inhibitors reduce the formation of oligomeric Aβ1–42 recognized by polyclonal A11 antibody, while negative control peptide does not. 10 μM Aβ1–42 was incubated alone (left-most column) or with 10-fold molar excess of each peptide-based inhibitor. Aliquots of the reaction were tested for antibody-binding at 6 hr, 24 hr, and 72 hr. 6E10 anti-amyloid beta (Tanzi, 2012; Hardy and Selkoe, 2002; Manczak and Reddy, 2014; Seward et al., 2013; Brier et al., 2016; Schwarz et al., 2016) was used as a loading control. Blots were quantified using ImageJ. (B) Unspliced dot blots from Figure 4C. Column key displayed at lower right is representative of all displayed blots in B.
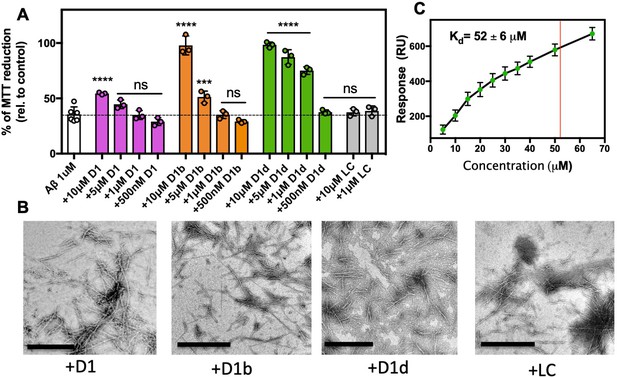
Inhibitors bind and block toxicity of aggregated Aβ1–42.
(A) The toxicity of already formed Aβ1–42 aggregates is lessened by peptide inhibitors. 10 μM Aβ1–42 was incubated alone for 12 hr at 37°C. Indicated molar ratio of inhibitor was added to the incubated Aβ1–42 and then diluted 1:10 with pre-plated N2a cells. Cytotoxicity was quantified using MTT dye reduction. Bars represent mean with individual technical replicates (n = 3–6; ns = not significant; ***, p<0.0005; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column). (B, C) Inhibitors bind to Aβ1–42 fibrils. (B) Peptide inhibitors do not disaggregate Aβ. 10 μM Aβ1–42 was incubated alone for 72 hr at 37 °C. Peptide inhibitors were added at 10-fold molar excess and incubated at RT for 24 hr before TEM analysis. Images were captured at 24,000x; scale bars are 500 nm. (C) Binding isotherm of inhibitor D1d to fibrillar Aβ1–42. The maximal response (RUmax) was derived by fitting sensorgrams obtained over a range of D1d concentrations to the binding model with a Kd of 52 ± 6 μM, displayed as a red line. These RUmax values are plotted (mean ± SD, n = 3) as a function of concentration and fitted to a one-to-one binding model, displayed as a black line.
-
Figure 4—source data 1
Toxicity data points—Aβ aggregates treated with inhibitors.
- https://doi.org/10.7554/eLife.46924.016
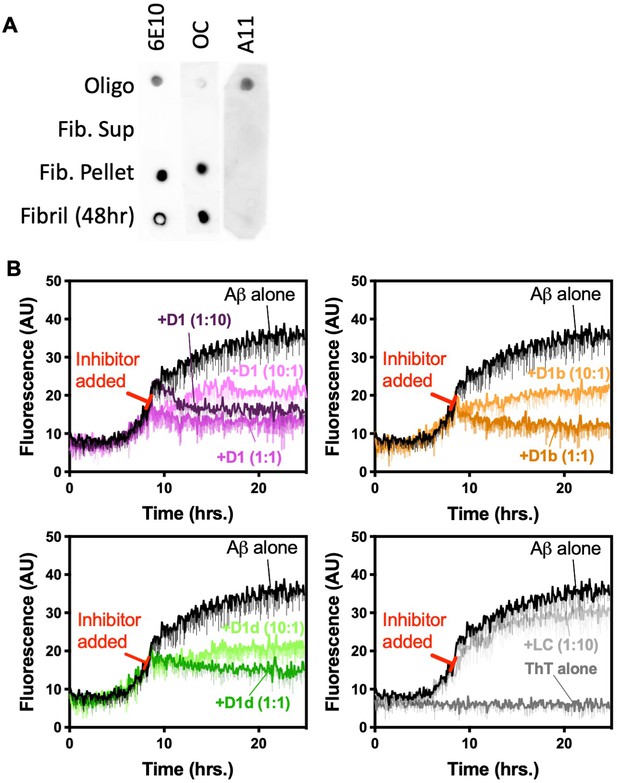
Aggregated Aβ conformations and inhibitor capping of Aβ fibrils.
(A) Conformation testing of Aβ preparations. 10 μM Aβ1–42 was incubated for 12 hr at 37°C in oligomer preparation, or 48 hr at 37°C with shaking for fibril conditions Fibrils were spun down at 50,000 rpm for 30 min. Samples were spotted on nitrocellulose membranes and probed with A11, OC and 6E10 antibodies. (B) Peptide inhibitors D1, D1b, and D1d reduce fibril elongation of Aβ1–42, while negative control peptide LC does not. 10 μM of Aβ1–42 was incubated alone until approximately half maximal ThT (19 A.U.) was observed under quiescent conditions at 37°C in polypropylene plates. Inhibitors were added at a 1:10, 1:1, or 10:1 molar ratio to Aβ1–42 reactions at 8.5 hr as indicated by red bar. Fibril formation was monitored using ThT fluorescence. Curves show the average of three technical replicates with one standard deviation below.
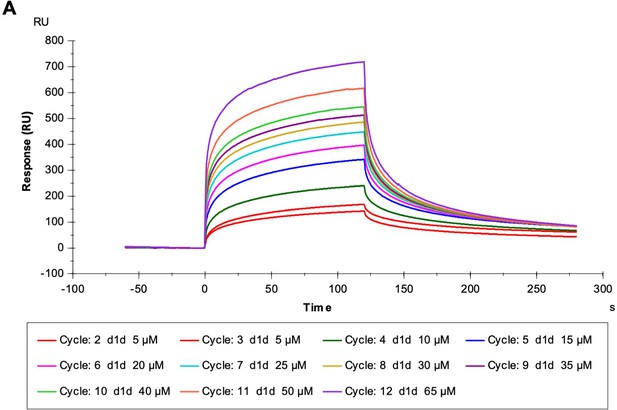
Representative Sensorgram obtained when D1d solutions at the indicated concentrations were flowed across the Aβ1–42 sensor chip.
The data of flow cell 1 (blank control) was subtracted from the data of flow cell 2.
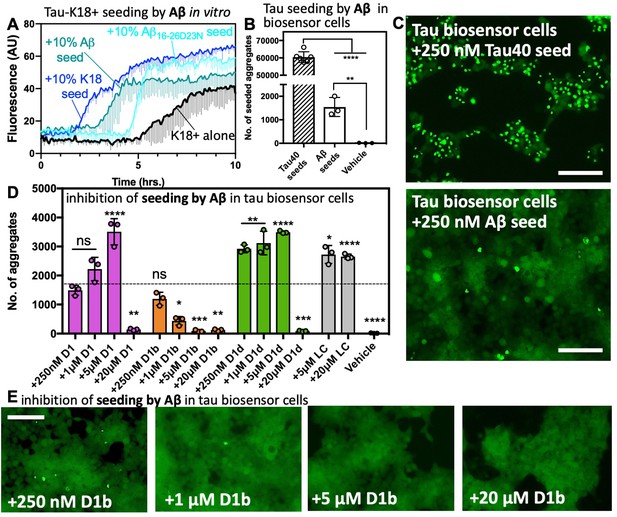
Tau aggregation is seeded by Aβ and reduced by structure-based inhibitors.
(A) 50 μM tau-K18+ was seeded with 10% monomer equivalent of pre-formed fibrils of Aβ1–42, Aβ16-26 D23N or tau-K18 under shaking conditions at 700 RPM at 37°C in PBS. Fibril formation was monitored using ThT fluorescence. Error bars below show the standard deviation of the average of three technical replicates. (B) The number of intracellular aggregates present in tau-K18CY biosensor cells normalized to cell confluence seeded by the addition of 250 nM tau40 or 250 nM Aβ1–42 fibrils. Error bars show the standard deviation of the mean of technical replicates (n = 3; ****, p=0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column, and **, p=0.0028 in unpaired t test of Aβ vs. vehicle) (C) Representative images of seeded cells from B at 10x magnification, scale bar 100 μm. (D and E). Concentration dependent inhibition of Aβ1–42 induced seeding of tau aggregation in tau-K18CY biosensor cells. (D) Average seeding by Aβ as a function of indicated inhibitor concentration. Error bars show the standard deviation of the mean of technical replicates (n = 3; ns = not significant; *, p<0.02; **, p<0.005; ***, p<0.001; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column), and the dotted line shows the mean number of aggregates from untreated Aβ1–42 fibrils. (E) Representative images of tau-K18CY biosensor cells showing the concentration-dependent effect of D1b on seeding. Cells are shown at 10X magnification, scale bar 100 μm.
-
Figure 5—source data 1
Tau biosensor seeding data points—Aβ and other amyloid fibrils with and without inhibtors.
- https://doi.org/10.7554/eLife.46924.020
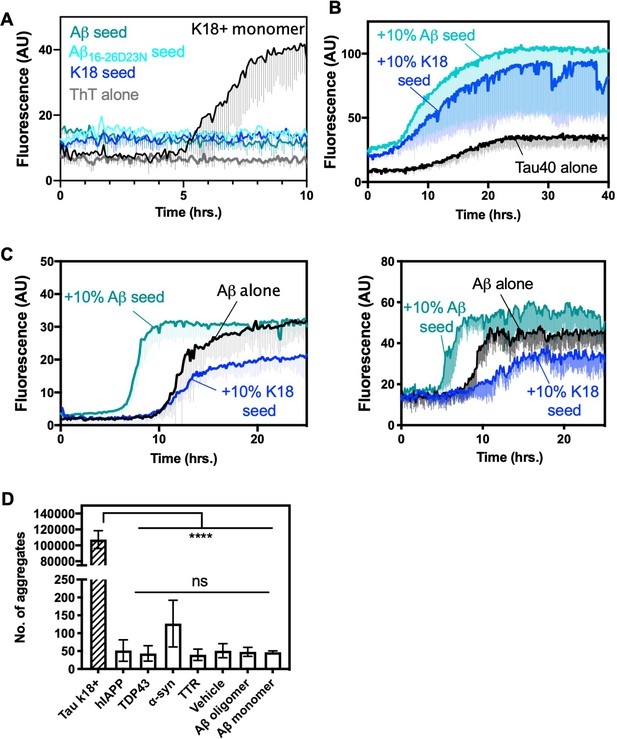
Extended ThT and seeding data.
(A) Extended ThT data of Tau seeding experiment. K18+ and Aβ1–42 seeds have some baseline ThT signal (B) 20 μM tau40 was seeded with 10% monomer equivalent of pre-formed fibrils of Aβ1–42 or tau-K18 under shaking conditions at 700 RPM at 37°C in buffer containing 0.5 mg/ml heparin. Fibril formation was monitored using ThT fluorescence. Aβ1–42 or tau-K18 seeds showed reduced lag times (9.4 and 10.2 hr, respectively) than unseeded tau40 (14.0 hr). Error bars show the standard deviation of the average of three technical replicates. T1/2 lag times were calculated using a four-parameter non-linear fit. (C) 10 μM Aβ1–42 was seeded with 10% monomer equivalent of pre-formed seed of Aβ1–42 or K18, under quiescent conditions at 37°C. Fibril formation was monitored using ThT fluorescence. Lines show average of three technical replicates. Left panel was performed in polystyrene plates, right panel was performed in polypropylene plates. (D) Tau-K18+ versus amyloid fibrils of other proteins, and non-fibrillar Aβ efficiency of seeding the HEK293 biosensor that stably expresses K18 yellow fluorescent protein (YFP) fusion. The cells were seeded with 250 nM final concentration of protein. Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3; ns = not significant; ****, p<0.0001 using an ordinary one-way ANOVA- Tukey’s relative to leftmost column or against all columns excluding leftmost, as indicated). Amyloid fibrils were confirmed by endpoint ThT or Electron Microscopy. Non-fibrillar Aβ was used fresh (monomeric) or incubated for 12 hr 37°C (oligomer).
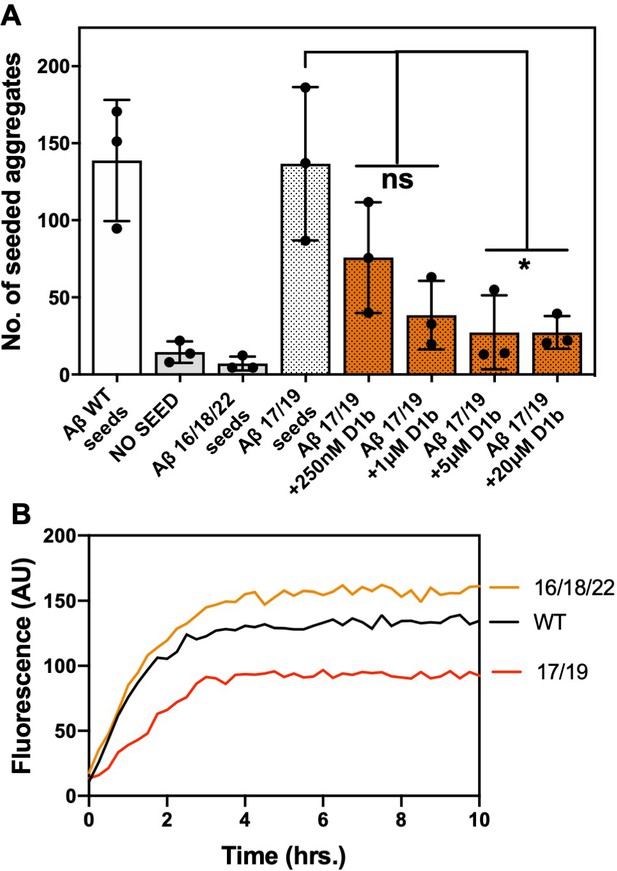
Seeding and inhibition of Aβ interface mutants.
(A) Aβ interface mutant constructs form fibrils. 25 μM of each Aβ constructs was incubated under quiescent conditions at 37°C. Fibril formation was monitored using ThT fluorescence. Lines show the average of three technical replicates. (B) Average seeding by Aβ mutant constructs and effect of indicated inhibitor concentration. Error bars show the standard deviation of the mean of technical replicates (n = 3; ns = not significant; *, p<0.04 using an ordinary one-way ANOVA- Dunnett’s relative to Aβ 17/19 seeds).
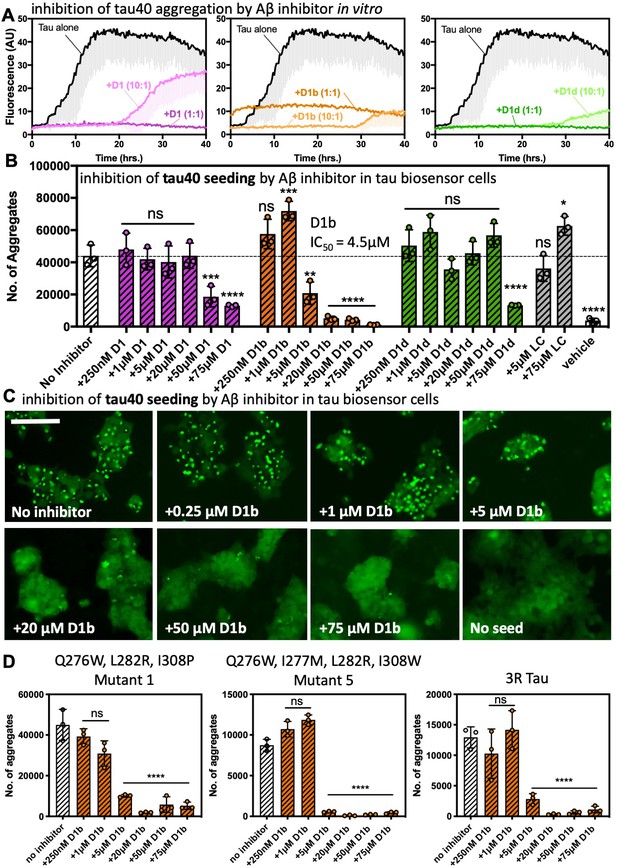
Aβ inhibitors also reduce fibril formation and seeding by tau40.
(A) Peptide inhibitors D1, D1b, and D1d reduce fibril formation of tau40. 10 μM tau40 monomer was incubated at a 1:10, 1:1, or 10:1 molar ratio to each inhibitor with 0.5 mg/ml heparin under shaking conditions at 700 RPM at 37°C. Fibril formation was monitored using ThT fluorescence. Plots show the average of three technical replicates with one standard deviation below. (B), (C). The effects of the inhibitors on seeding by tau40 fibrils in tau-K18CY biosensor cells. The cells were seeded with 250 nM tau40 fiber (final concentration); in samples with inhibitor, tau40 fibers were incubated with indicated final concentrations of peptide inhibitor for one hour prior to addition to cells. (B) Average number of aggregates at the indicated inhibitor concentrations, Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3; ns = not significant; *, p<0.03; **, p<0.023; ***, p<0.0008; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column). dotted line represents number of aggregates from untreated tau40 fibrils. IC50 value was calculated from the dose–response plot of inhibitor D1b. (C). Representative images of effect of D1b on seeding. Cells are shown at 10X magnification, scale bar 100 μm. (D) Seeding from tau interface mutation fibrils in tau-K18CY biosensor cells is reduced by D1b. Experiment was performed as above. Average number of aggregates at the indicated inhibitor concentrations, Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3; ns = not significant; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column).
-
Figure 6—source data 1
Tau biosensor seeding data points—tau and tau interface mutations with and without inhibitor.
- https://doi.org/10.7554/eLife.46924.026
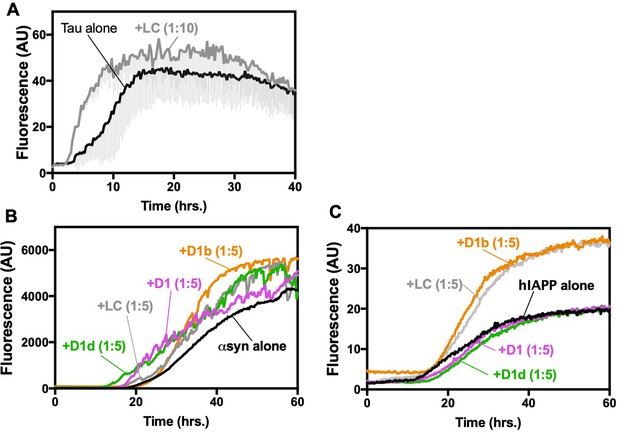
Specificy of inhibitors.
(A) Control peptide inhibitor LC does not reduce fibril formation of tau40. 10 μM of tau40 was incubated at a 1:10 molar ratio to LC with 0.5 mg/ml heparin under shaking conditions at 700 RPM at 37°C. Fibril formation was monitored using ThT fluorescence. Lines show the average of three technical replicates with one standard deviation below. (B, C) Designed inhibitors are not general amyloid inhibitors. (B) 50 μM of α-synuclein was incubated at a 1:5 molar ratio to peptide inhibitors under shaking conditions at 700 RPM at 37°C. Lines show the average of three technical replicates. (C) 10 μM hIAPP was incubated at a 1:5 molar ratio to peptide inhibitors under quiescent conditions at 37°C. Lines show the average of three technical replicates.
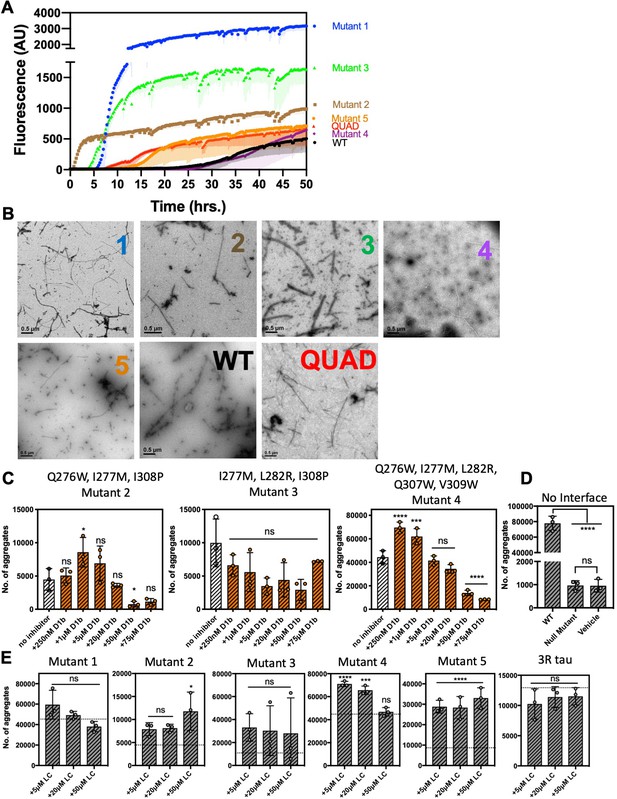
Extended tau40 interface mutation data.
(A, B) Tau 40 interface mutant constructs form fibrils. (A) 10 μM of each tau40 constructs was incubated with 0.5 mg/ml heparin under shaking conditions at 700 RPM at 37°C. Fibril formation was monitored using ThT fluorescence. Lines show the average of three technical replicates. (B) Negative-stain TEM analysis confirms the results of the ThT assays in (A). Scale bars are 0.5 µm. (C, D, E) Extended data of seeding by tau interface mutation fibrils in tau-K18CY biosensor cells. The cells were seeded with 250 nM tau40 fiber (final concentration); in samples with inhibitor, tau40 fibers were incubated with indicated final concentrations of peptide inhibitor for one hour prior to addition to cells. Average number of aggregates at the indicated inhibitor concentrations, Bars represent mean with individual technical replicates, error bars display one standard deviation (n = 3; ns = not significant; *, p<0.05; ***, p<0.0005; ****, p<0.0001 0001 using an ordinary one-way ANOVA- Dunnett’s in (C, E) relative to leftmost column (C) or dotted line (E). Panel D was calculated using ANOVA-Tukey’s, relative to leftmost column and between Null mutant and vehicle columns). (C) Interface mutations less affected by D1b. (D) Construct containing all interface mutations is seeding incompetent. (E) Seeding interface mutants are not reduced by control inhibitor LC. Extended ANOVA data included as a supplementary file.
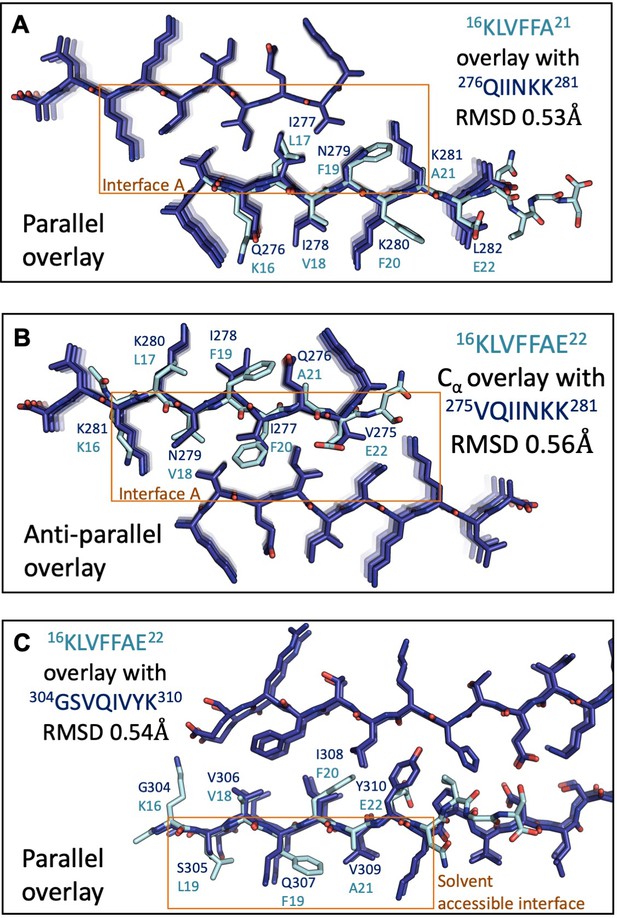
The spines of Aβ 16–26 D23N and tau are structurally similar.
(A, B) Aβ 16–26 D23N overlay with tau 274–283 in parallel and antiparallel orientations. (A) 32 backbone atoms differ from each other by 0.53 Å RMSD. RMSD values were calculated using LSQ in Coot. (B) The Cα atoms differ from each other by 0.56 Å RMSD. Backbone and side chain rotamers were optimized with Foldit over 2000 iterations to minimize energy to −603 REU. (C) Aβ 16–22 overlay with tau 304–310 in parallel, backbone atoms differ from each other by 0.54 Å RMSD. RMSD values were calculated using LSQ in Coot.
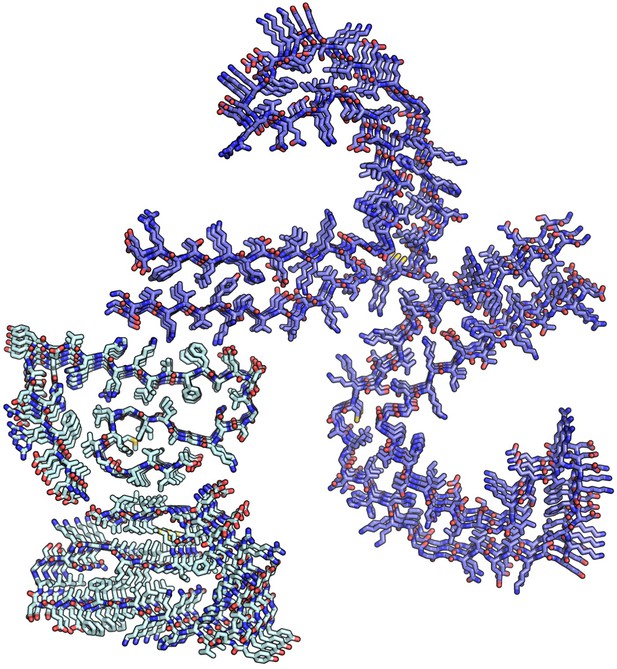
Hetero-seeding model from side of Aβ fibril.
Residues 16–21 of Aβ (5OQV) were superimposed on 304–309 of Tau (6HRF). Tau was then manually moved perpendicular to the fibril axis to make a complementary surface with 5OQV. Backbone and side chain rotamers were optimized with Foldit to minimize energy to −1517REU.
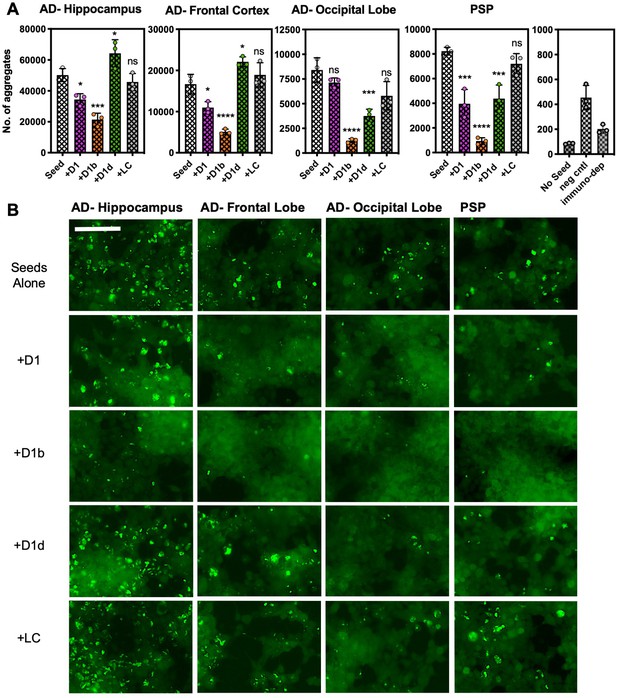
Peptide inhibitors reduce seeding by crude brain-extract from tauopathy donor tissue.
Brain lysate was prepared in TBS buffer from three brain regions of one AD patient, and from a one sample of a PSP patient lacking Aβ plaques. Brain lysate from a non-disease patient (neg cntl) and a tau immunodepleted sample from PSP tissue are in right panel. Cells were seeded with a 1/400 dilution of brain tissue lysate; for samples with inhibitor, lysates were incubated with inhibitor overnight prior to addition to cells. A concentration of 10 µM peptide was used for all of the experiments shown. (A) The average number of aggregates seeded by lysate from each respective brain region, with or without addition of inhibitors. Bars represent mean with individual technical replicates (n = 3; ns = not significant; *, p<0.05; ***, p<0.0005; ****, p<0.0001 using an ordinary one-way ANOVA- Dunnett’s relative to leftmost column). (B) Representative images of seeded biosensor cells from A shown at 10X magnification, scale bar 100 μm. Extended ANOVA data included as a supplementary file.
-
Figure 7—source data 1
Tau biosensor seeding data points—brain lysate with and without inhibitor.
- https://doi.org/10.7554/eLife.46924.028
Tables
Statistics of MicroED data collection and atomic refinement.
https://doi.org/10.7554/eLife.46924.005| KLVFFAENVGS | |
|---|---|
| Excitation Voltage (kV) | 200 |
| Electron Source | field emission gun |
| Wavelength (Å) | 0.0251 |
| Total dose per crystal (e-/ Å2) | 2.7 |
| Frame rate (frame/s) | 0.3–0.5 |
| Rotation rate (°/s) | 0.3 |
| #crystals used | 13 |
| Total angular rotation collected (°) | 941 |
| Merging Statistics | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 11.67, 51.91, 12.76 |
| α, β, γ (°) | 90, 114.18, 90 |
| Resolution (Å) | 11.64–1.4 (1.44–1.40)* |
| Rmerge | 24.0% (65.2%) |
| No. Reflections | 47,598 (1966) |
| Unique Reflections | 2355 (163) |
| Completeness (%) | 86.2% (78.0%) |
| Multiplicity | 21 (12) |
| I/σ | 9.06 (2.88) |
| CC1/2 | 99.5% (69.7%) |
| Refinement Statistics | |
| No. reflections | 2354 |
| Reflections in test set | 236 |
| Rwork | 23.7% |
| Rfree | 28.3% |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.5 |
| Avg. B factor (Å2) | 9.46 |
| Wilson B factor (Å2) | 7.2 |
| Ramachandran (%) | |
| Favored | 100% |
| Allowed | 0% |
| Outliers | 0 |
-
*Highest resolution shell shown in parenthesis.
| Reagent type | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | HEK293 -K18 (P301S)-EYFP | Diamond Laboratory | ||
| Cell line (M. musculus) | Neuro-2a cell line | ATCC | Cat # CCL-131 RRID:CVCL_0470 | |
| Antibody | Goat anti-Mouse IgG H and L (FITC) secondary antibody | Abcam | Cat# ab7064, RRID:AB_955234 | WB 1:10000 |
| Antibody | Donkey anti-Rabbit IgG H and L (FITC) secondary antibody | Abcam | RRID:AB_955259 | WB 1:10000 |
| Antibody | A11 (rabbit polyclonal) | Millipore | Cat# AB9234 RRID:AB_11214948 | WB 1:500 |
| Antibody | OC (rabbit polyclonal) | Millipore | Cat# AB2286 RRID:AB_1977024 | WB 1:2000 |
| Antibody | 6E10 (mouse monoclonal) | BioLegend | Cat# 803003, RRID:AB_2564652 | WB 1:5000 |
| Antibody | A11-09 mOC 64 mOC 24 mOC 116 mOC 104(rabbit monoclonal) | Glabe Laboratory, Abcam | WB 1:100 | |
| Peptide, recombinant protein | Recombinant Amyloid Beta 1–42 (pET15b-MBP-AB) | This paper | See Materials and methods section | |
| Peptide, recombinant protein | (D)-LYIWVQ | Genscript | >95% purity | |
| Peptide, recombinant protein | (D)-LYIWIWRT | Genscript | >95% purity | |
| Peptide, recombinant protein | (D)-LYIWIQKT | Genscript | >95% purity | |
| Peptide, recombinant protein | (L)-LYIWVQ | Genscript | >95% purity | |
| Peptide, recombinant protein | Recombinant Tau40 (1–441, pET22b) | This paper | See Materials and methods section | |
| Peptide, recombinant protein | Recombinant K18(244-372) K18+ (244-380) (PNG2) | This paper | See Materials and methods section | |
| Peptide, recombinant protein | KLVFFAENVGS | Genscript | >98% purity | |
| Chemical compound, drug | 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) dye | Sigma | Cat# M5655 | |
| Chemical compound, drug | ThioflavinT | Sigma | CAS ID: 2390-54-7 | |
| Software, algorithm | XDS | http://xds.mpimf-heidelberg.mpg.de/ | RRID:SCR_015652 | |
| Software, algorithm | CCP4 | http://www.ccp4.ac.uk/ | RRID:SCR_007255 | |
| Software, algorithm | PHENIX | PMID: 20124702 | http://www.phenixonline.org/; RRID: SCR_014224 | Model building and refinement |
| Software, algorithm | Coot | PMID: 20383002 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; RRID: SCR_014222 | Model building |
| Software, algorithm | Rosetta | https://www.rosettacommons.org/home | RRID:SCR_015701 | |
| Software, algorithm | Graphpad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 8 |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | Foldit | http://fold.it/ | RRID:SCR_003788 |
Additional files
-
Source data 1
Extended ANOVA data.
- https://doi.org/10.7554/eLife.46924.029
-
Supplementary file 1
Computed binding properties of designed inhibitors to Aβ 16KLVFFAEN23.
- https://doi.org/10.7554/eLife.46924.030
-
Transparent reporting form
- https://doi.org/10.7554/eLife.46924.031





