A requirement of Polo-like kinase 1 in murine embryonic myogenesis and adult muscle regeneration
Figures
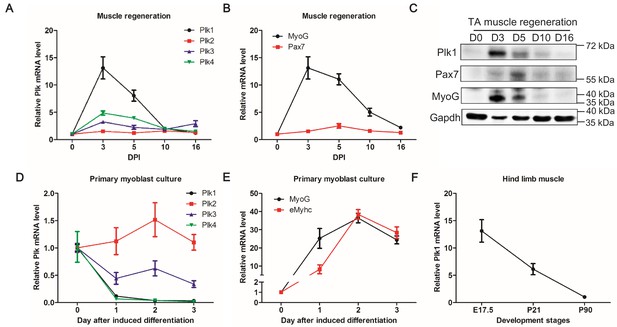
Expression patterns of Plks during muscle regeneration and differentiation.
(A–B) Relative mRNA levels of Plks and myogenic factors Pax7 and MyoG in TA muscles from mice (n = 4) at various timepoints after CTX induced injury, determined by qPCR, DPI: Days post injury; (C) Representative protein level of Plk1, Pax7 and Myog at various timepoints during muscle regeneration; (D–E) qPCR showing relative mRNA levels of Plks and myogenic differentiation markers (Myogenin and eMhyhc: embryonic myosin heavy chain) at various timepoints of primary myoblast differentiation (n = 3, biological samples); (F) qPCR analysis of Plk1 expression in TA muscles from 17.5 day, 3 week and 3-month-old mice (n = 3, biological samples).
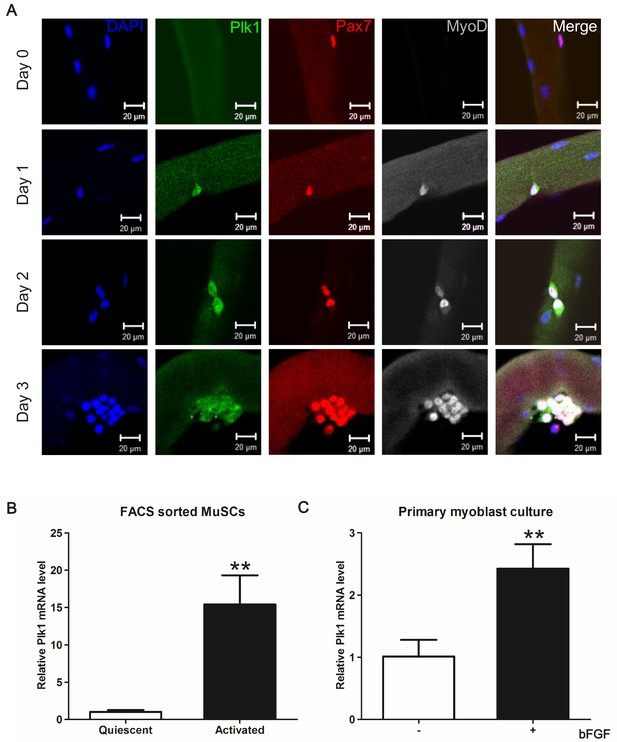
Plk1 is specifically expressed in activated MuSCs.
(A) Plk1 immunofluorescence in Pax7+ MuSCs attached on freshly isolated EDL myofibers (Day 0) or after cultured for 1–3 days from three male wildtype mice (at least 10 fibers were collected at each time point, quantification of MyoD+Plk1+ MuSCs were based on 200 cells on myofibers cultured for 72 hr), scale bar: 10 μm; (B) Relative levels of Plk1 mRNA in quiescent and activated satellite cells, data represent mean ± s.e.m. (t-test: **p<0.01; n = 3, biological replicates); (C) Relative levels of Plk1 mRNA in primary myoblast treated with (+) or without (-) 4 ng/ml bFGF for 24 hr, data represent mean ± s.e.m. (t-test: *p<0.05; n = 4, biological replicates).
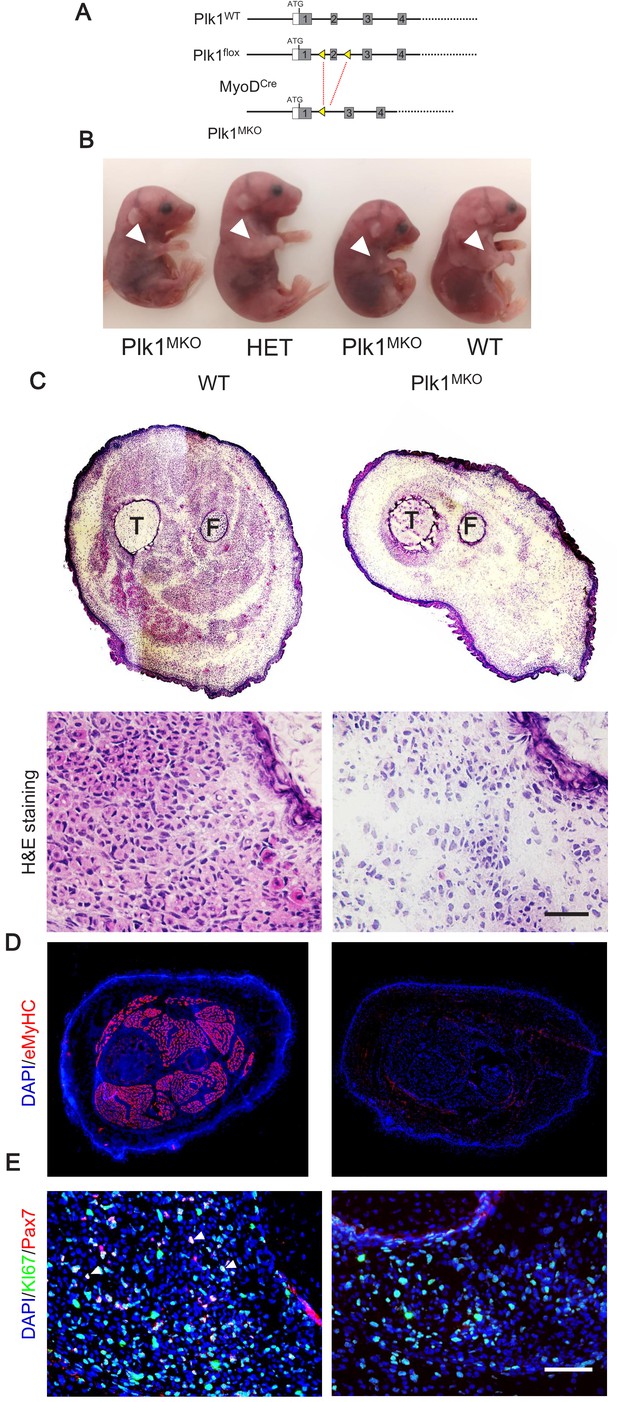
Loss of Plk1 in myogenic progenitors leads to embryonic lethality.
(A) Targeting strategy for myogenic progenitor specific deletion of Plk1, boxes represent exons and triangles represent LoxP; (B) Representative images of WT, heterozygous KO (Het) and Plk1MKO embryos at stage E16.5, arrowheads points to the forelimbs, showing the; (C) H and E staining of E16.5 hindlimb cross-sections (upper panel) and magnified representative area (bottom panel, scale bar: 50 μm) showing lack of muscle fibers (labeled by eosin in pink) in Plk1MKO embryos, T: Tibia, F: Fibula; (D) Immunofluorescence of eMyHC (marking myofibers) in E16.5 whole limb cross-sections; (E) Immunofluorescence of Pax7 and KI67 in limb cross-sections of embryos at E16.5, scale bar: 50 μm.
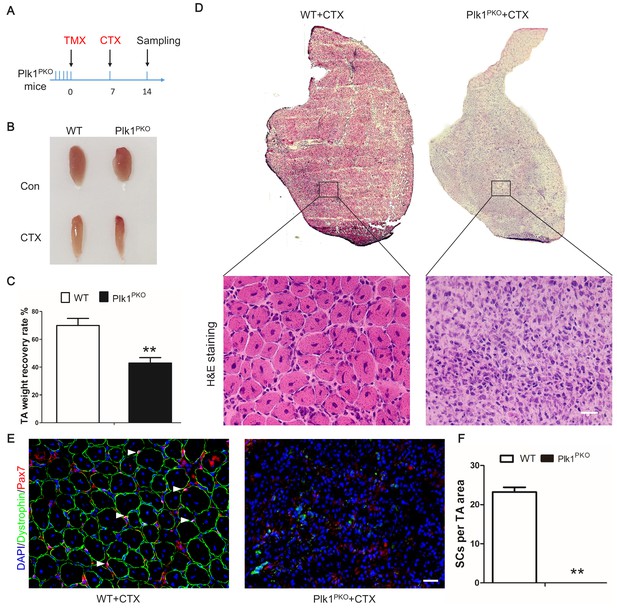
Plk1 deletion in MuSCs impairs muscle regeneration in vivo.
(A) Experimental design for tamoxifen (TMX, four 8-week-old male mice from each group received a daily intraperitoneal injection for 5 consecutive days) induced deletion of Plk1 in Pax7CreER::Plk1f/f (Plk1PKO) mice, following by cardiotoxin (CTX) injection to induce muscle degeneration and regeneration. Numbers indicate timing of TMX induction (5 consecutive days of injection following by 7 days of chasing), CTX injection and sample collection (7 days after injury); (B) Representative images of TA muscles in WT and Plk1PKO mice; (C) TA muscle weight recovery at 7 days after CTX injury, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4, biological replicates, mice were males at around 11 weeks old when sampled); (D) H and E staining of TA muscle cross-sections (upper panel) and magnified representative areas at 7 days after CTX injury (bottom panels, scale bar: 20 μm); (E) Immunofluorescence of Pax7 and Dystrophin (to outline myofibers) in TA muscle cross-sections at 7 days after CTX injury, scale bar: 20 μm; (F) The average number of MuSCs per microscopic area, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4, biological replicates, all MuSCs from five radom areas were counted for each mouse).
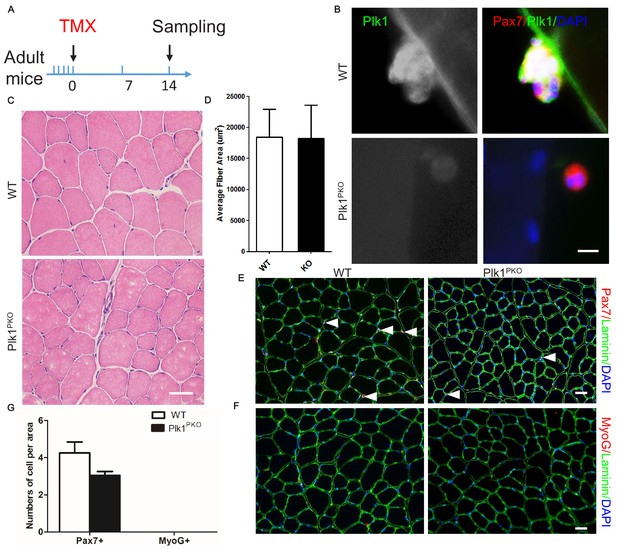
Plk1 deletion in MuSCs have minimal impact on resting muscle.
(A) Schematics showing timing of TMX induction and sample collection (four 8-week-old male mice from either group received a daily intraperitoneal injection of TMX for 5 consecutive days, then sampled after 14 days); (B) Immunofluorescence of Plk1 (green), Pax7 (red) and DAPI (blue) in MuSCs attached on EDL myofibers after cultured for 3 days, scale bar: 10 μm; (C) H and E staining of TA muscle cross-sections after 14 days of TMX induction; (D) The average fiber area of TA muscle cross-sections after 14 days of TMX induction, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4, biological replicates, all fibers from five random areas were counted for each mouse); (E) Immunofluorescence of Pax7 (red) and Laminin (green) in TA muscle cross-sections 7 days after TMX induction, scale bar: 20 μm, white arrowheads point to Pax7+DAPI+ MuSCs; (F) Immunofluorescence of MyoG (red) and Laminin (green) in TA muscle cross-sections 14 days after TMX induction, scale bar: 20 μm; (G) The average number of Pax7+ or MyoG+ cells per microscopic area, data represent mean ± s.e.m. (t-test: **p<0.01; n = 6 biological replicates, all MuSCs from five random areas were counted for each mouse).
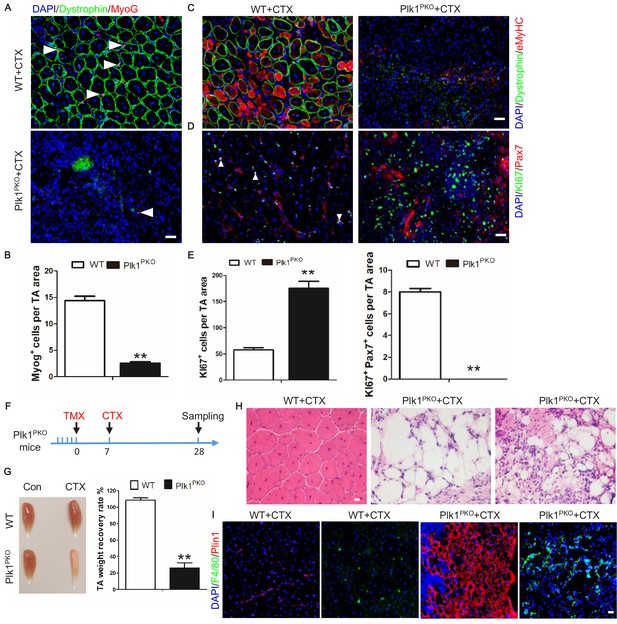
Plk1 deletion in MuSCs impairs muscle regeneration in vivo.
(A) Immunofluorescence of MyoG (red) and Dystrophin (green) in TA muscle cross-sections 7 days after CTX injury, scale bar: 20 μm, white arrows point to myogenic cells; (B) The average number of MyoG+ cells per microscopic area, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4 biological replicates, all MyoG+ cells from five random cross-sectional areas were counted for each mouse); (C) Immunofluorescence of eMyHC (red) and Dystrophin (green) in TA muscle cross-sections 7 days after CTX injury, scale bar: 20 μm; (D) Immunofluorescence of KI67 (green) and Pax7 (red) in TA muscle cross-sections 7 days after CTX injury, scale bar: 20 μm; (E) The average number of KI67+ cells and KI67+Pax7+ cells per microscopic area, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4 biological replicates, all KI67+ cells from five random cross-sectional areas were counted for each mouse); (F) Experimental design for tamoxifen (TMX, three 3-month-old male mice in each group received a daily intraperitoneal injection for 5 consecutive days) induced deletion of Plk1 in Plk1PKO mice, following by cardiotoxin (CTX) injection to induce muscle degeneration and regeneration. Numbers indicate timing of TMX induction (following by 7 days of chasing), CTX injection and sample collection (in days, sampling at 21 days after CTX injury); (G) Representative images of TA muscles in WT and Plk1PKO mice at 21 days after CTX injury (left panel), TA muscle weight recovery at 21 days after CTX injury (right panel), data represent mean ± s.e.m. (t-test: **p<0.01; n = 3, biological replicates); (H) H and E staining of TA muscle cross-sections at 21 days after CTX injury, scale bar: 20 μm; (I) Immunofluorescence of F4/80 and Perilipin-1 (to outline adipocytes) in TA muscle cross-sections at 21 days after CTX injury, scale bar: 20 μm.
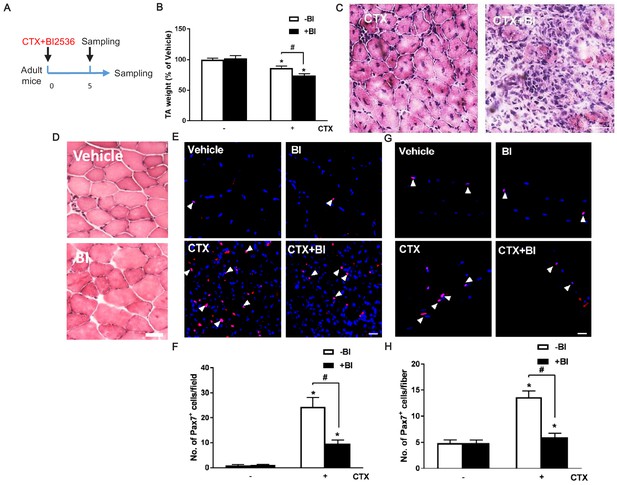
Plk1 inhibition by BI2623 impairs muscle regeneration.
(A) Schematic showing timing of BI2623 and CTX injection, and sample collection (Samples were collected 5 days after 50 μl saline or 50 μl of 10 μM CTX injection in the absence or presence of 10 μg BI2536, from four 2-month-old male mice); (B) TA weight at 5 days post-CTX injection in the absence or presence of BI, data represent mean ± s.e.m. (t-test: *p<0.05; n = 4, biological replicates); (C–D) Representative H and E staining of injured or non-injured TA muscles at 5 days post-CTX injection in the absence or presence of BI2536 (BI), red fibers with central nuclei represent regenerating myofibers, scale bar: 50 μm; (E) Representative images of Pax7 (red) staining in muscle cross section at 5 days post-CTX injection in the absence or presence of BI, nuclei were counterstained with DAPI (blue), scale bar: 20 μm; (F) Quantification of Pax7+ cells per field in muscle cross-section, co-staining of Pax7 and DAPI were recognized as Pax7+ cells, indicated by white arrows, data represent mean ± s.e.m. (t-test: */#p<0.05; n = 4, biological replicates, all MuSCs from five random cross-sectional areas were counted for each mouse); (G) Representative images of Pax7 (red) staining in isolated EDL single fibers from 5 days post CTX injection in the absence or presence of BI, nuclei were counterstained with DAPI (blue), scale bar: 20 μm; (H) Quantification of Pax7+ cells per single fiber, at least 20 fibers were counted for each treatment data represent mean ± s.e.m. (t-test: */#p<0.05; n = 4, biological replicates, all MuSCs from five random images were counted for each mouse).
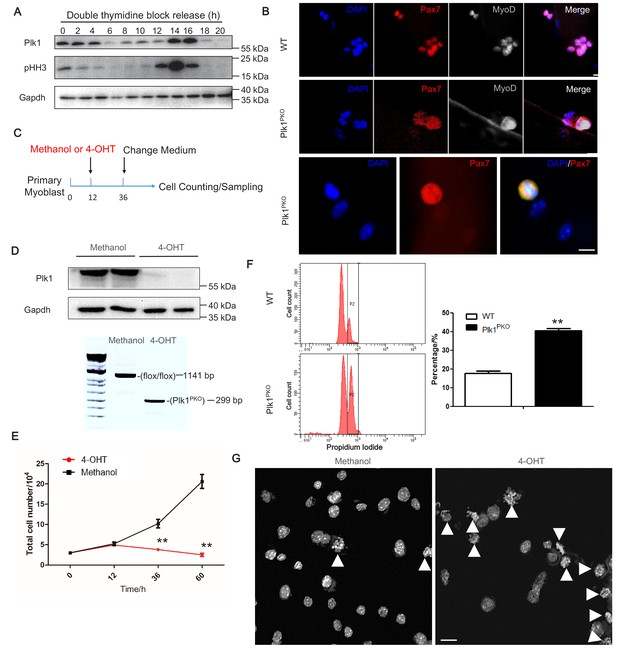
Plk1 deletion in MuSCs leads to cell cycle arrest.
(A) Western blot showing relative protein levels of Plk1 and cell cycle marker pHH3 at different time points after WT myoblast were synchronized by double thymidine block (DTB) protocol and released; (B) Immunofluorescence of Pax7 and MyoD in MuSCs on cultured EDL myofibers (72 hr) isolated from WT and Plk1PKO mice 7 days after TMX induced deletion of Plk1, scale bar: 10 μm, (at least 10 fibers were collected from four individual 11-week-old male mice); (C) Schematics showing 4-hydroxy-tamoxifen (4-OHT) induced deletion of Plk1 in primary myoblasts isolated from Plk1PKO mice (Primary myoblasts were isolated from four 6-week-old male mice, and frozen at −80°C and stored for the following experiments), Methanol treatment is the vehicle control; (D) Western blot showing effective knockout of Plk1 after 4-OHT induction (upper panel) and PCR analysis of genomic DNA showing the DNA recombination 4-OHT induction; (E) Quantification of the average numbers of myoblast per well, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4, biological replicates); (F) Flow cytometry analysis of methanol or 4-OHT treated myoblast with propidium iodide staining to mark DNA content (left panel), and quantification of percentage of myoblasts containing double-DNA content (right panel), data represent mean ± s.e.m. (t-test: **p<0.01; n = 3, biological replicates); (G) DAPI staining of myoblasts at 30 hr after methanol or 4-OHT induction to reveal DNA morphology, scale bar: 10 μm, white arrowheads indicated cells containing unsegregated chromosomes.
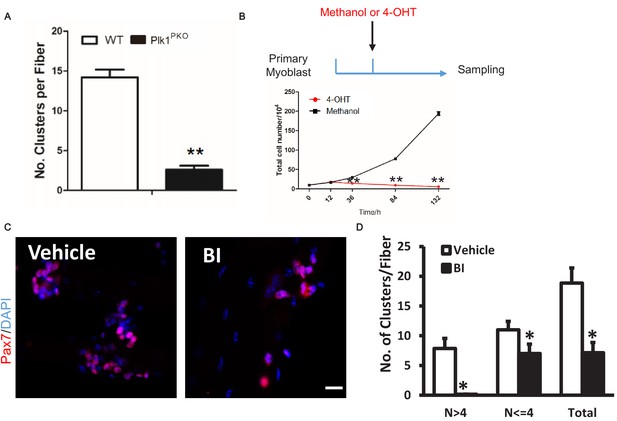
Plk1 deletion in MuSCs leads to cell cycle arrest.
(A) Quantification of MuSCs on 3 day cultured EDL myofibers isolated from WT and Plk1PKO mice 14 days after TMX induction, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4, biological replicates, MuSCs from at least 10 fibers were counted for each mouse); (B) Total myoblast numbers after 0, 12, 36, 84 and 132 hr methanol or 4-OHT induction, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4 biological replicates, cell numbers were counted from three replicates from four WT or Plk1PKO myoblasts); (C) Immunofluorescence of Pax7 and DAPI in MuSCs on 3 day cultured EDL myofibers isolated from BI or vehicle treated mice, scale bar: 10 μm; (D) Quantification of (C), data represent mean ± s.e.m. (t-test: *p<0.05; n = 4, biological replicates, MuSCs from at least 10 fibers were counted for each mouse).
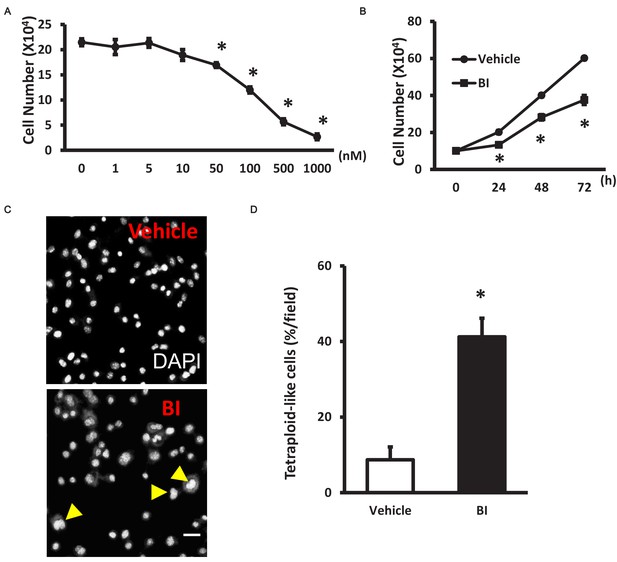
Plk1 inhibition in myoblast leads to cell cycle arrest.
(A) Total myoblast numbers after 24 hr treatment of 0, 1, 5, 10, 50, 100, 500, 1000 nM BI or (B) 24, 48, 72 hr treatment of 100 nM BI, data represent mean ± s.e.m. (t-test: *p<0.05; n = 4, biological replicates, cell numbers were counted from three replicates from four vehicle or BI-treated myoblasts); (C) Representative images of primary myoblast morphology 24 hr after treatment of 100 nM BI, nuclei were stained with DAPI (white), scale bar: 20 μm; (D) Quantification of tetraploid-like cells, data represent mean ± s.e.m. (t-test: *p<0.05; n = 5, biological replicates, 25–50 myoblasts/replicate, three replicates from five different time points were counted for quantification).
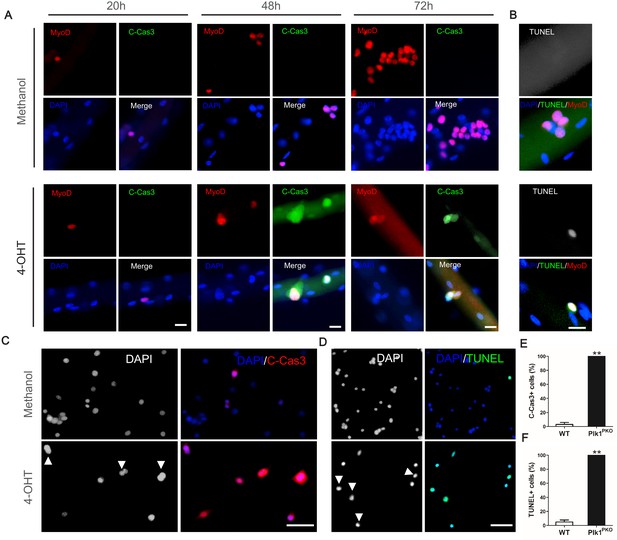
Plk1 deletion leads to apoptosis of MuSCs.
(A) Representative images of MyoD (red) and cleaved Caspase-3 (green) staining in MuSCs located on EDL myofibers cultured for 20, 48 and 72 hr, myofibers were isolated from Plk1PKO mice and treated with 4-OHT to induce deletion of Plk1 at time 0, Methanol is the vehicle control, scale bar: 10 μm, at least 10 cells from 10 single myofibers per mouse (n = 3 mice) were counted; (B) Immunofluorescence of Myod (red) and TUNEL (green) in MuSCs on EDL myofibers cultured for 42 hr, scale bar, 10 μm, at least 10 cells from 10 single myofibers per mouse (n = 3 mice) were counted; (C–D) Representative images of cleaved Caspase-3 (C) and TUNEL (D) staining in primary myoblasts at 48 hr after methanol or 4-OHT treatment, nuclei were counterstained with DAPI, arrowheads indicate cells arrested at M-phase, scale bar: 50 μm; (E–F) Quantification of percentages of Caspase-3+ (E) and TUNEL+ (F) cells, data represent mean ± s.e.m. (t-test: **p<0.01; n = 3, biological replicates (mice), three replicates/mouse, 25–50 myoblasts/replicate were analyzed).
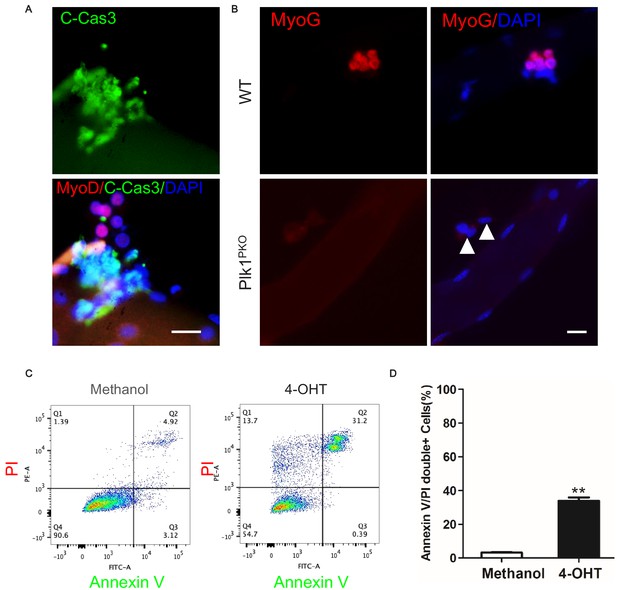
Deletion of Plk1-null MuSCs is due to apoptosis but not differentiation.
(A) Representative images of MyoD (red) and cleaved Caspase-3 (green) staining in 72 hr cultured MuSCs of fresh isolated EDL single fibers treated with H2O2 for 30 min, scale bar: 10 μm; (B) Immunofluorescence of Myog (red) and DAPI (blue) in MuSCs on 3 day cultured EDL myofibers isolated from WT and Plk1PKO mice 7 days after TMX induction, white arrowheads point to undivided MuSCs on fibers from Plk1PKO mice, (C) Flow cytometry analysis of methanol or 4-OHT treated myoblast with Annexin V and PI staining to mark apoptotic cells; (D) Quantification of percentage of Annexin V/PI double positive myoblasts, data represent mean ± s.e.m. (t-test: **p<0.01; n = 4, biological replicates).
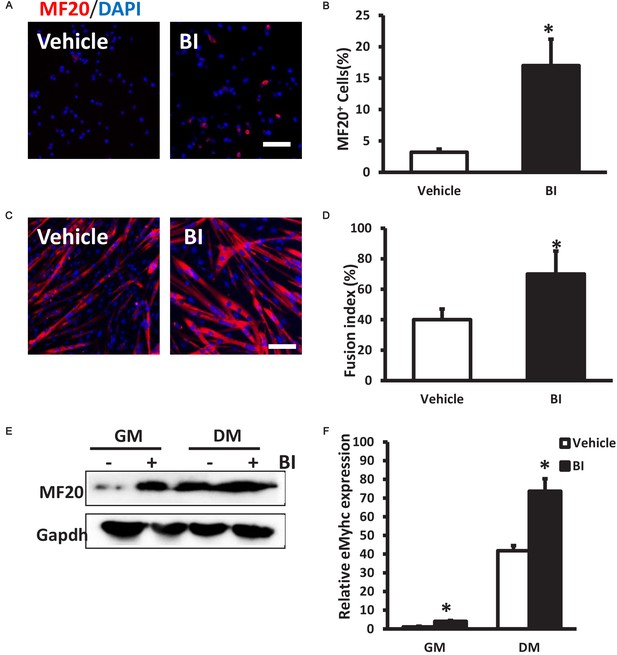
Inhibition of Plk1 leads to premature differentiation of primary myoblasts.
(A) Representative images of MF20 staining (red) in primary myoblast 48 hr after 100 nM BI treatment in growth medium (GM, DMEM +20%FBS +4 ng/μl bFGF), nuclei were counterstained with DAPI (blue); (B) Quantification of MF20+ cells per field in BI2536 treated myoblast, five fields were counted for each treatment, data represent mean ± s.e.m. (t-test: *p<0.05; n = 3, biological replicates); (C) Representative images of MF20 staining in primary myoblast 48 hr after BI treatment in differentiation media (DM, DMEM +2% HS), nuclei were counterstained with DAPI; (D) Fusion index of differentiated myoblasts, five fields were counted for each treatment, data represent mean ± s.e.m. (t-test: *p<0.05; n = 3, biological replicates), scale bar: 50 μm; (E) Representative protein level of MF20 in primary myoblast 48 hr after BI treatment in GM and DM (n = 3); (F) Relative mRNA level of eMyHC in primary myoblast after BI treatment in GM and DM, data represent mean ± s.e.m. (t-test: *p<0.05; n = 3, biological replicates).
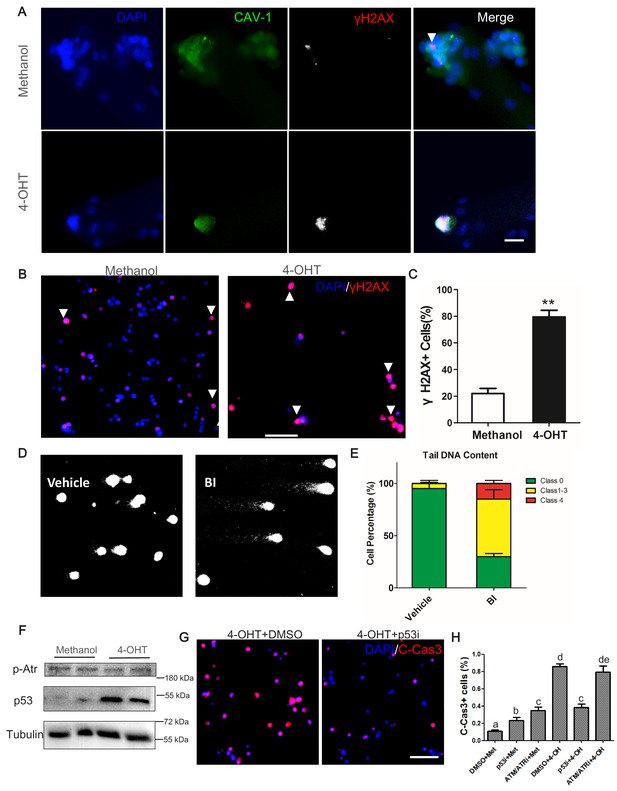
Plk1-null MuSCs excessively accumulate markers of DNA damage response and undergo p53-mediated apoptosis.
(A) Immunofluorescence of γH2AX (red) and Caveolin-1 (green) in MuSCs on EDL myofibers cultured for 72 hr. Myofibers were isolated from Plk1PKO mice and treated with 4-OHT to induced Plk1 deletion (methanol: vehicle control), scale bar: 10 μm; (B) Representative images of γH2AX staining in primary myoblast isolated from Plk1PKO mice, treated with 4-OHT or methanol, and cultured for 48 hr, nuclei were counterstained with DAPI, scale bar: 50 μm; (C) Quantification of percentages γH2AX+ myoblasts after treatment shown in B, data represent mean ± s.e.m. (t-test: **p<0.01; n = 3, biological replicates(mice), three replicates/mouse, 25–50 myoblasts/replicate were analyzed); (D) Representative images of single-cell gel electrophoresis under alkaline conditions in primary myoblasts 24 hr after BI2536 treatment; (E) Percentage of myoblasts with different levels of DNA damage, presented as no damage, moderate damage (classes 1–3) and maximal damage (class 4), data represent mean ± s.e.m. (t-test: *p<0.05; n = 3, biological replicates (mice), three replicates/mouse, 25–50 myoblasts/replicate were analyzed); (F) Western blot showing relative levels of p53 and Phospho-ATR in Plk1PKO primary myoblast at 24 hr after methanol or 4-OHT treatment; (G) Representative images of cleaved Caspase-3 (red) staining in primary myoblast 24 hr after 4-OHT together with DMSO (left panel) or p53 inhibitor (p53i) treatment (right panel), nuclei were counterstained with DAPI, scale bar: 50 μm, (H) Quantification of C-Cas3+ apoptotic cells at 24 hr after methanol or 4-OHT together with p53 inhibitor or ATM/ATR inhibitor treatment, data represent mean ± s.e.m. (t-test: p<0.05; n = 3, biological replicates, 25–50 myoblasts from three replicates from three individual mice were counted for quantification).
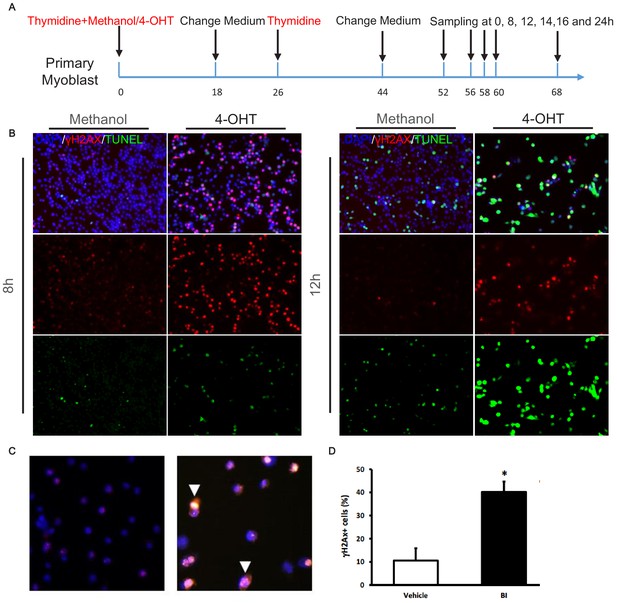
Plk1-null MuSCs undergo DNA damage response induced apoptosis.
(A) Schematic showing timing of double thymidine and 4-OHT/methanol treatment, and sample collection; (B) Immunofluorescence of γH2AX and TUNEL after double thymidine block/release for 8 and 12 hr, (C) Representative images of γH2AX staining in primary myoblasts 24 hr after BI treatment; (D) Percentage of γH2AX + myoblasts after treatment, data represent mean ± s.e.m. (t-test: *p<0.05; n = 5, biological replicates, 25–50 myoblasts from three replicates from five different treatments were counted for quantification).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Plk1flox | Pubmed | PMID: 27417127 | Dr. Guillermo de Cárcer (Spanish National Cancer Research Centre) |
| Genetic reagent (M. musculus) | Myodcre | Jackson Laboratory | Stock #: 014140 RRID:IMSR_JAX:014140 | |
| Genetic reagent (M. musculus) | Pax7creER | Jackson Laboratory | Stock #: 012476 RRID:IMSR_JAX:012476 | |
| Antibody | Pax7(PAX7) mouse monoclonal | Developmental Studies Hybridoma Bank | Cat# pax7 RRID:AB_528428 | IHC (1:10) |
| Antibody | MyoG mouse monoclonal | Developmental Studies Hybridoma Bank | Cat# PCRP-MYOG-1C5 RRID:AB_2722260 | IHC (1:500) WB (1:1000) |
| Antibody | MF20 mouse monoclonal | Developmental Studies Hybridoma Bank | Cat# MF 20 RRID:AB_2147781 | IHC (1:50) WB (1:200) |
| Antibody | eMyHC mouse monoclonal | Developmental Studies Hybridoma Bank | Cat# F1.652 RRID:AB_528358 | IHC (1:100) |
| Antibody | Plk1 rabbit polyclonal | Cell Signaling | Cat# 4535 RRID:AB_2252687 | IHC (1:500) WB (1:2000) |
| Antibody | MyoD mouse monoclonal | Santa Cruz Biotechnology | Cat# sc-377460 | IHC (1:300) |
| Antibody | Dystrophin rabbit polyclonal | Abcam | Cat# ab15277 RRID:AB_301813 | IHC (1:1000) |
| Antibody | Ki67 rabbit polyclonal | Abcam | Cat# ab15580 RRID:AB_443209 | IHC (1:1000) |
| Antibody | Phospho-Histone H3 rabbit polyclonal | Cell Signaling Technology | Cat# 9701 RRID:AB_331535 | WB (1:1000) |
| Antibody | Phospho-ATR rabbit monoclonal | Cell Signaling Technology | Cat# 30632 RRID:AB_2798992 | WB (1:1000) |
| Antibody | p53 rabbit polyclonal | Cell Signaling Technology | Cat# 9282 RRID:AB_331476 | WB (1:1000) |
| Antibody | Cleaved Caspase-3 rabbit polyclonal | Cell Signaling Technology | Cat# 9661 RRID:AB_2341188 | IHC (1:500) |
| Antibody | Anti-H2A.X, phospho mouse monoclonal | Abcam | Cat# ab26350 RRID:AB_470861 | IHC (1:1000) |
| Antibody | 488 goat polyclonal antimouse IgG1 | Thermo Fisher Scientific | Cat# A-21121 RRID:AB_141514 | IHC (1:1000) |
| Antibody | 488 goat polyclonal antirabbit IgG | Thermo Fisher Scientific | Cat# A-11034 RRID:AB_2576217 | IHC (1:1000) |
| Antibody | 568 goat polyclonal antimouse IgG1 | Thermo Fisher Scientific | Cat# A-21124 RRID:AB_2535766 | IHC (1:1000) |
| Antibody | 488 goat polyclonal antirabbit IgG | Thermo Fisher Scientific | Cat# A-21244 RRID:AB_2535812 | IHC (1:1000) |
| Antibody | HRP-conjugated goat polyclonal anti-rabbit IgG | Jackson ImmunoResearch Labs | Cat# 111-035-003 RRID:AB_2313567 | WB (1:10000) |
| Antibody | HRP-conjugated goat polyclonal anti-mouse IgG | Jackson ImmunoResearch Labs | Cat# 115-035-003 RRID:AB_10015289 | WB (1:10000) |
| Chemical compound, drug | BI2536 | Selleckchem | Cat# S1109 | |
| Chemical compound, drug | Pifithrin-α | Sigma | Cat# P4359 | |
| Chemical compound, drug | CGK733 | Sigma | Cat# C9867 | |
| Chemical compound, drug | propidium iodide | Sigma | Cat# P4170 | |
| Chemical compound, drug | Annexin V | Invitrogen | Cat# V13241 |
Additional files
-
Supplementary file 1
Genotypes and distribution of Plk1 conditional knockout embryos.
- https://doi.org/10.7554/eLife.47097.018
-
Supplementary file 2
Primers used in this study.
- https://doi.org/10.7554/eLife.47097.019
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47097.020





