HDX-MS reveals structural determinants for RORγ hyperactivation by synthetic agonists
Figures
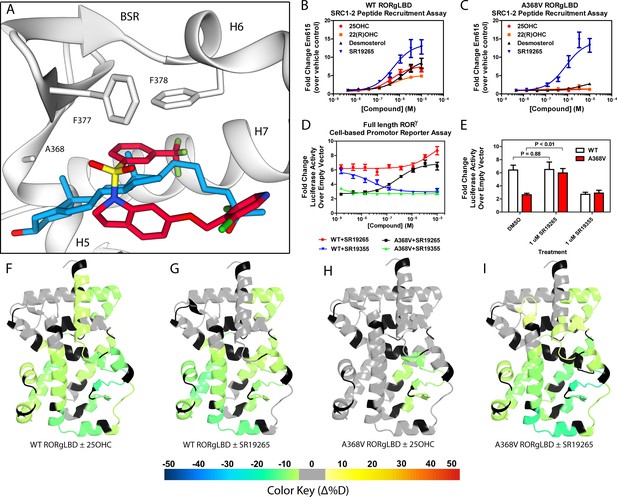
Ligand binding is required to activate RORγ in vitro.
(A) The binding poses of SR19265 (red) and 25-hydroxycholesterol (blue, 25OHC) are compared. Both compounds are shown in the context of co-crystal structure solutions with RORγ (PDB ID 3L0L) where helices 5, 6, and 7 (H5, H6, and H7) as well as the beta sheet region (BSR) are shown. SR19265 and 25OHC are shown in red and blue respectively. A368V was found to be a mutation that selectively disrupts endogenous ligand binding presumably through steric clashes. WT and A368V RORγLBD were tested in an AlphaScreen-based SRC1-2 coactivator peptide recruitment assay in panels B and (C). (D) HEK293T cells were transiently transfected with a vector encoding full length RORγ and a 5xRORE-luciferase reporter. The WT and A368V variant were tested for their baseline activity as well as their response to SR19265 and SR19355. (E) Summary of luciferase assay activity showing A368V is a loss of function mutation and that activity is recovered with SR19265. Activity from WT and A368V variant RORγ are shown as white and red bars, respectively. (F-I) Solvent exchange kinetics of the WT (panels F and G) and A368V (panels H and I) variant RORγ LBDs were assessed using differential HDX-MS to compare 25OHC (panels F and H) and SR19265 (panels G and I) treated protein to vehicle control treated protein. The change in percent deuterium uptake of 54 peptides were averaged and consolidated for each amino acid and overlaid onto the RORγ LBD in the active conformation (PDB ID 3L0L). Gray and black regions of the protein indicate no significant changes to exchange and no sequence coverage, respectively.
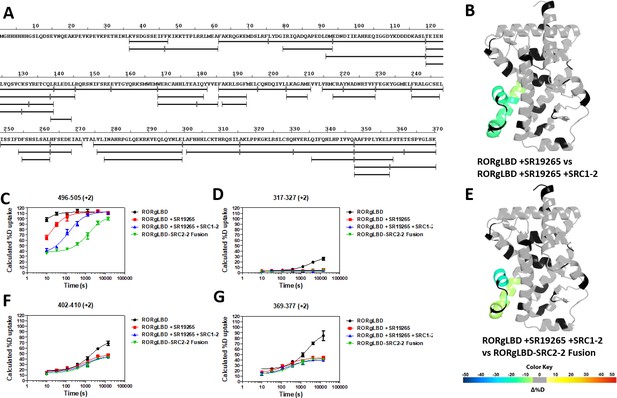
Overview of HDX-MS characterization of RORγ (related to Figures 1–5).
(A) Sequence coverage map of peptic RORγLBD peptides. The sequence of the HisSUMO-RORγLBD construct is shown above and observed peptides are shown as black sticks. Selected deuterium build up curves of peptides from H12, H3, H7, and the BSR in panels C,D,F, and G, respectively. The numbering scheme for peptides is adjusted to match the sequence numbering of RORγ. (B and E) Selected differential HDX-MS experiments painted onto RORγLBD in the active conformation (PDB ID 3L0L). Differential of RORγLBD liganded with SR19254 ±SRC1-2 peptide is shown in B. The differential of RORγLBD + SR19265+SRC1-2 vs RORγLBD-SRC2 fusion is shown in E.
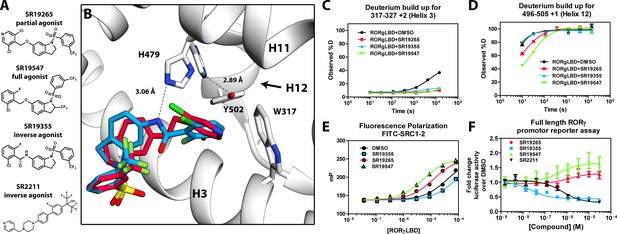
Mechanism of action for select N-arylsulfindoline RORγ modulators.
(A) Representative structures of four modulators and their pharmacology. (B) Co-crystal structure solutions of SR19265 and SR19355 reveal mechanism of action. A peptide bond on SR19355 acts as a hydrogen bond donor for H479 and disrupts H479 and Y502 hydrogen bond. (C, D) Differential HDX-MS was employed to evaluate ligand dependent structural perturbations in solution. Representative deuterium build-up plots showing Helix 3 (C, H3) and Helix 12 (D, H12) dynamics. (E) fluorescence polarization results showing gradient of affinity for coactivator peptide depending on compound. (F) RORγ modulators were tested in a cell-based promotor reporter assay. Vectors encoding full length RORγ and a 5xRORE-Luciferase promotor reporter were transiently transfected into HEK293T cells. These cells were seeded into a 384 well plate and tested for dose-dependent responses to compounds.
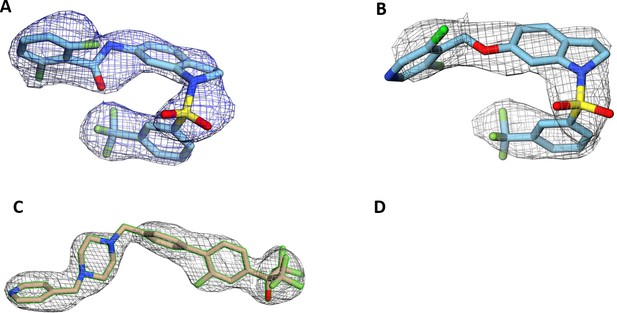
Electron density maps are shown for compounds of interest (related to Figures 1 and 2).
2Fo-Fc densities are contoured at 1.5 σ. SR19265 and SR19355 are shown in panels (A and B). SR2211 in chain A and chain B are shown in figures (C and D).
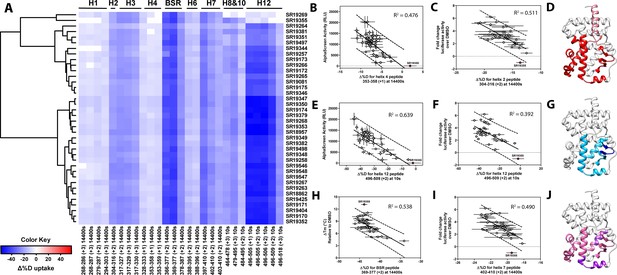
Differential HDX-MS and activity screening of 38 RORγ modulators reveals structural determinants for activation.
(A) Δ%D (difference from DMSO treated control) values for 38 RORγ modulators are plotted as a heat map according to the color key at the bottom left. Each row represents a compound and each column represents a peptide indicated by the labels at the right and bottom respectively. The locations of each peptide in the crystal structure are labeled along the top. A hierarchical clustering algorithm based on Ward’s method was performed to cluster compounds based on their exchange signatures and the corresponding dendrogram is shown on the left. (B-J) Activity correlation analysis results found that structural dynamics measurements correlate with functional activity. Regions of RORγ that correlate with coactivator peptide recruitment, thermal stability, and activation in cell-based assays are shown in panel D, G, and J, respectively. Lighter shades indicate that the slope of a linear regression was significantly (adjusted p value < 0.05) non-zero, while darker shades indicate an R (Takeda et al., 2012) was greater than 0.4. Representative coactivator peptide recruitment correlation plots for helix 12 and helix four are shown in B and E, respectively. The correlation between thermal stability and the BSR peptide are shown in panel (H). Representative correlation plots cell-based activity are shown for H2, H12 and H7 are shown in panels C, F, and I, respectively.
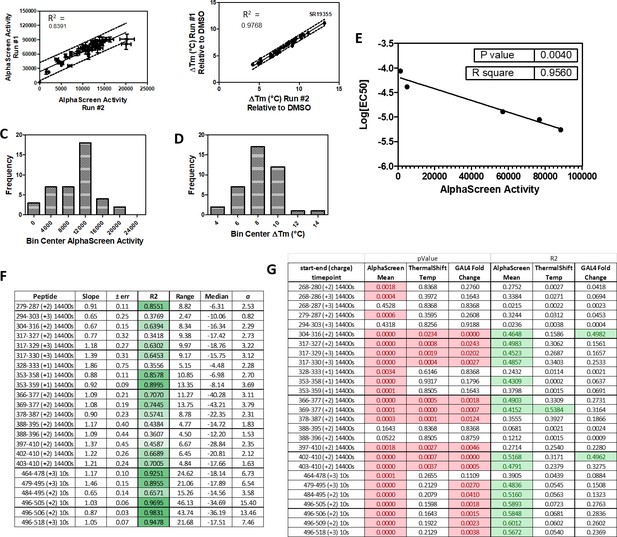
Summary and reproducibility of biochemical and two timepoint HDX-MS screening data (related to Figures 3 and 4).
38 RORγ modulators were tested in AlphaScreen-based peptide recruitment assays and thermal shift assays in two separate preparations of compounds and protein to assess reproducibility. The run to run comparisons are shown in panel (A and B). The distributions of the compound activities are shown in panels (C and D). AlphaScreen-based peptide recruitment activity correlated with fluorescent polarization-based peptide recruitment EC50 values as shown in panel (E). The Reproducibility of 2 timepoint HDX-MS screening was performed using two batches of protein and the results of the run-to-run correlations are shown in panel (F).
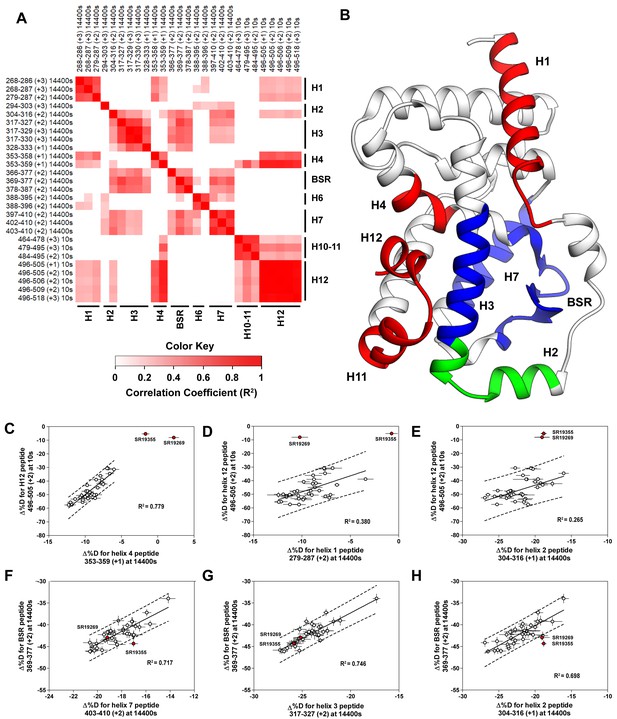
Covariation analysis of two timepoint HDX-MS screening reveals concerted ligand-dependent changes in protein dynamics.
(A) Correlogram showing correlation coefficients (R2) of Δ%D values between peptides. Peptides are labeled as start-end (charge) timepoint across the top and left sides of the panel while the location of the peptides are labeled along the bottom and right sides of the panel. Correlation coefficients that are significantly non-zero (adjusted P values < 0.01) slope are colored. (B) Perturbation values generally correlated in three groups. Group one correlated with H12 and consists of H1 and H4 and is colored red. Group two correlates with the BSR consists of H7 and H3 and is colored Blue. A peptide spanning residues 304–316 is shown in green. (C-H) Representative correlation plots showing distributions of compound perturbation values across peptides indicated by the x- and y-axis. Linear regression models are shown as solid black lines and the 95% confidence interval for prediction is shown as dashed lines. SR19355 and SR19269 (indicated in red) were often found as outliers for models involving helix 12 (panels C-E).
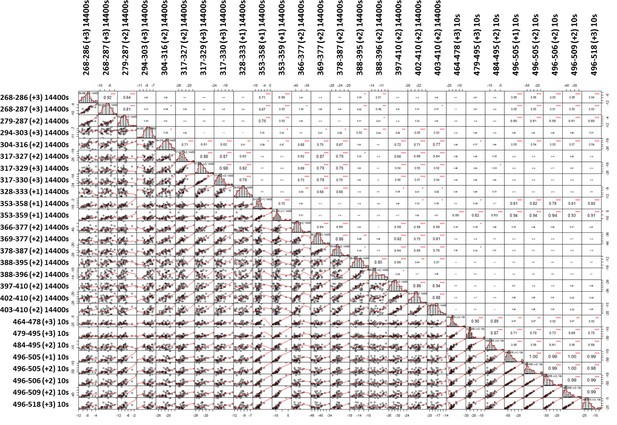
A weighted correlation analysis of the two timepoint compound screening dataset shows widespread correlation between peptides.
Across the diagonal are histograms showing normal distributions of Δ%D for each peptide. Below the diagonal are correlation plots comparing peptide perturbation values of the peptides above and on the left of the corresponding plot. Above the diagonal are correlation coefficients (pearson’s R value) of a linear model describing the variances of peptide perturbation values of the peptides below and left of the corresponding R value. Significance is indicated for adjusted P vales <0.1, 0.05, 0.01 and 0.001 as. , *,**, and *** respectively.
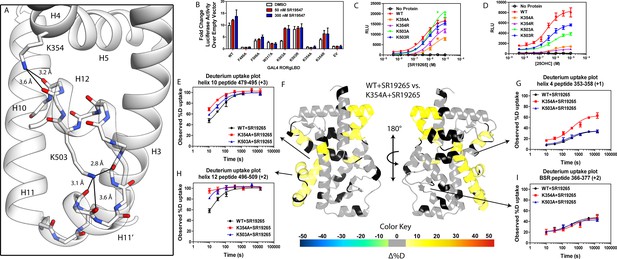
intramolecular interactions are necessary for activation by endogenous agonists.
(A) RORγ in the active conformation (PDB ID 3L0L) reveals that K354 and K503 form several hydrogen bonds with backbone carbonyl shown in red. The functional consequences of these interactions in stabilizing the active conformation were assessed with site directed mutagenesis, promotor-reporter assays, (B), coactivator peptide recruitment assays (C and D) and HDX-MS (E-I). (B) RORγ variants were tested in a GAL4 chimera promoter reporter assay using a dual-glo assay format. SR19265 and 25OHC were tested for dose-dependent responses in AlphaScreen-based SRC1-2 coactivator peptide recruitment assay in panels C and D respectively. Solvent exchange kinetics of WT and mutant RORγLBDs bound to SR19265 were compared using HDX-MS in panels E-I. Deuterium uptake plots for helix 10, 4, 12 and the BSR are shown in panels E, G, H, and I, respectively. Results from differential HDX-MS comparing WT + SR19265 and K354A + SR19265 were consolidated and painted onto the structure of RORγ (PDB ID 3L0L) in panel G.
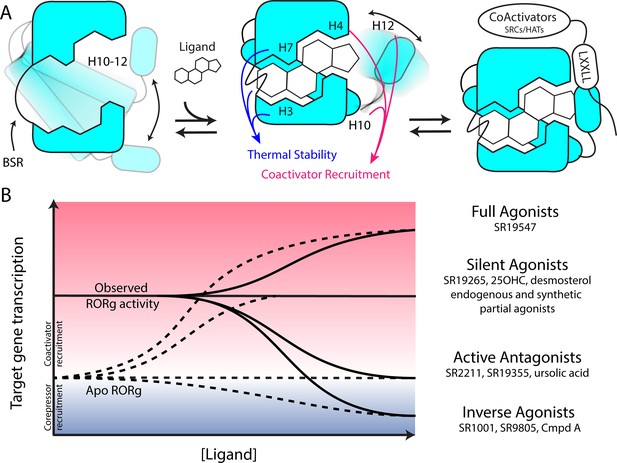
Model for ligand-dependent activation of RORγ.
(A) Apo RORγ exhibits extensive conformational dynamics in the absence of compound. Ligand recognition through the BSR, H3 and H7 confer thermal stability and allows for H10-12 stability where H12 and H4 dynamics are the key structural determinants for coactivator affinity and receptor hyperactivation. (B) Based on mutagenesis studies, apo RORγ is presumed to be inactive. The observed high basal activity is likely due to activation of RORγ by endogenous agonists. Compounds that outcompete these endogenous ligands and activate RORγ to the observed levels should be considered silent agonists.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (HEK293T) | HEK293T | ATCC | Cat:CRL-11268 | |
| Transfected construct (Human) | pBIND-RORγHinge-LBD | Kumar et al., 2010 DOI:10.1124/mol.109.060905 | Addgene_128091 | |
| Transfected construct (Human) | pGL4.35[luc2P/9XGAL4 UAS/Hygro] Vector | Promega | Cat:E1370 | |
| Transfected construct (Human) | pSPORTV6-hRORγ | Kumar et al., 2012 DOI:10.1021/cb200496y | Addgene_128093 | |
| Transfected construct (Human) | pGLA4.1-5xRORE- Luciferase | Kumar et al., 2012 DOI:10.1021/cb200496y | Addgene_128094 | |
| Commercial assay or kit | Britelite Luciferase assay reagent | Perkin Elmer | Cat:6066766 | |
| Commercial assay or kit | Dual-Glo Luciferase assay reagents | Promega | Cat:E2920 | |
| Recombinant DNA reagent | pESUMO-RORγLBD | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | Addgene_128090 | |
| Recombinant DNA reagent | pESUMO-RORγLBD-SRC2 | This Paper | Addgene_128089 | |
| Commercial assay or kit | Q5 mutagenesis kit | New England Biolabs | Cat:E0554S | |
| Sequence-based reagent | Q5_hRORC_F378A_Sense | Sigma | N/A | CACGGTCTTTgcgGAAGGCAAATAC |
| Sequence-based reagent | Q5_hRORC_F378A_Antisense | Sigma | N/A | CGGTTGTCAGCATTGTAG |
| Sequence-based reagent | Q5_hRORC_F388A_Sense | Sigma | N/A | CATGGAGCTGgcgCGAGCCTTGG |
| Sequence-based reagent | Q5_hRORC_F388A_Antisense | Sigma | N/A | CCACCGTATTTGCCTTCA |
| Sequence-based reagent | Q5_hRORC_F486A_Sense | Sigma | N/A | GCTGCAGATCgcgCAGCACCTCC |
| Sequence-based reagent | Q5_hRORC_F486A_Antisense | Sigma | N/A | CTTTCCACATGCTGGCTAC |
| Sequence-based reagent | Q5_hRORC_K503A_Sense | Sigma | N/A | TCCACTCTACgcgGAGCTCTTCAGCACTG |
| Sequence-based reagent | Q5_hRORC_K503A_Antisense | Sigma | N/A | GGGAAAGCGGCTTGGACC |
| Sequence-based reagent | Q5_hRORC_K503R_Sense | Sigma | N/A | TCCACTCTACcgtGAGCTCTTCAGCACTGAAAC |
| Sequence-based reagent | Q5_hRORC_K503R_Antisense | Sigma | N/A | GGGAAAGCGGCTTGGACC |
| Sequence-based reagent | Q5_hRORC_F506A_Sense | Sigma | N/A | CAAGGAGCTCgcgAGCACTGAAACC |
| Sequence-based reagent | Q5_hRORC_F506A_Antisense | Sigma | N/A | TAGAGTGGAGGGAAAGCG |
| Sequence-based reagent | Q5_hRORC_Y502F_Sense | Sigma | N/A | caagGAGCTCTTCAGCACTGAAACC |
| Sequence-based reagent | Q5_hRORC_Y502F_Antisense | Sigma | N/A | aagagTGGAGGGAAAGCGGCTTG |
| Sequence-based reagent | Q5_hRORC_A368V_Sense | Sigma | N/A | GATGTGCCGGgtgTACAATGCTGAC |
| Sequence-based reagent | Q5_hRORC_A368V_Antisense | Sigma | N/A | CTAACCAGCACCACTTCC |
| Sequence-based reagent | Q5_hRORC_K354A_Sense | Sigma | N/A | TGTGCTTCTCgccGCAGGAGCAATG |
| Sequence-based reagent | Q5_hRORC_K354A_Antisense | Sigma | N/A | ATCTGGTCATTCTGGCAG |
| Sequence-based reagent | Q5_hRORC_K354R_Sense | Sigma | N/A | TGTGCTTCTCcggGCAGGAGCAATG |
| Sequence-based reagent | Q5_hRORC_K354R_Antisense | Sigma | N/A | ATCTGGTCATTCTGGCAG |
| Peptide, recombinant protein | Biotin-Linker-SRC1-2 | Lifetein | N/A | Biotin-Ahx-SPSSHSSLTERHKILHRLLQEGSP |
| Peptide, recombinant protein | HisSUMO-hRORgLBD(265-518) | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | N/A | |
| Peptide, recombinant protein | RORgLBD(265-507) | This Paper | N/A | For co-crystallography with SR2211 |
| Peptide, recombinant protein | RORgLBD(265-507)-linker-SRC2-2 | This Paper | N/A | For co-crystallography with SR19265 and SR19355 |
| Commercial assay or kit | AlphaLISA Anti-HIS-Acceptor beads | Perkin Elmer | Cat:AL128C | |
| Commercial assay or kit | AlphaScreen Streptavidin-Donor beads | Perkin Elmer | Cat:6760002S | |
| Chemical compound, drug | Fc1cccc(Cl)c1OCc2ccc3ccn(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR18862 | |
| Chemical compound, drug | Fc1cccc(Cl)c1OCc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR18957 | |
| Chemical compound, drug | Fc1cccc(Cl)c1CCc2 ccc3ccn(c3c2)S(=O)(=O) c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19081 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19170 | |
| Chemical compound, drug | FC(F)(F)c1cccc(c1)S(=O)(=O)N2CCc3ccc(OCc4c(Cl)cccc4Cl)cc23 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19171 | |
| Chemical compound, drug | Cc1cccc(Br)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19172 | |
| Chemical compound, drug | Cc1cccc(C)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19173 | |
| Chemical compound, drug | COc1ccccc1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19174 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3ccn(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19257 | |
| Chemical compound, drug | Cc1cccc(c1)S(=O) (=O)N2CCc3ccc(OCc4c(F)cccc4Cl)cc23 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19258 | |
| Chemical compound, drug | COc1cccc(c1)S(=O)(=O)N2CCc3ccc(OCc4c(F)cccc4Cl)cc23 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19263 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cc(Cl)cc(Cl)c4 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19264 | |
| Chemical compound, drug | FC(F)(F)c1cccc(c1)S(=O)(=O)N2CCc3ccc(OCc4c(Cl)cncc4Cl)cc23 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19265 | |
| Chemical compound, drug | COc1cccnc1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19266 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cncc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19267 | |
| Chemical compound, drug | FC(F)(F)Oc1ccccc1COc2ccc3CCN(c3c2) S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19268 | |
| Chemical compound, drug | CC(Oc1ccc2CCN(c2c1)S(=O)(=O)c3cccc(c3)C(F)(F)F)c4c(F)cccc4Cl | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19269 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(n4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19344 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(Cl)c4 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19346 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cc(ccn4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19348 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4ccnc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19349 | |
| Chemical compound, drug | COc1cc(Cl)ccc1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19350 | |
| Chemical compound, drug | COc1c(Cl)cccc1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19351 | |
| Chemical compound, drug | FC(F)(F)c1cccc(c1)S(=O)(=O)N2CCc3ccc(OCc4c(Cl) ccnc4Cl)cc23 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19352 | |
| Chemical compound, drug | COc1ccc(Cl)cc1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19353 | |
| Chemical compound, drug | Fc1cccc(Cl)c1C(=O)Nc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19355 | |
| Chemical compound, drug | CN(C(=O)c1c(F)cccc1Cl)c2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19379 | |
| Chemical compound, drug | Fc1cccc(Cl)c1CNc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19381 | |
| Chemical compound, drug | CN(Cc1c(F)cccc1Cl)c2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19382 | |
| Chemical compound, drug | CC1Cc2ccc(OCc3c(F)cccc3Cl)cc2N1S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19425 | |
| Chemical compound, drug | Fc1cncc(Cl)c1COc2ccc3CCN(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19497 | |
| Chemical compound, drug | FC(F)(F)c1cccc(c1)S(=O)(=O)N2CCc3ccc(OCc4c(Cl)cncc4C(F)(F)F)cc23 | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19498 | |
| Chemical compound, drug | FC(F)(F)C1Cc2ccc(OCc3c(Cl)cncc3Cl)cc2N1S(=O)(=O)c4cccc(c4)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19546 | |
| Chemical compound, drug | Fc1cccc(Cl)c1COc2ccc3CC(N(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19547 | |
| Chemical compound, drug | Fc1cncc(Cl)c1COc2ccc3CC(N(c3c2)S(=O)(=O)c4cccc(c4)C(F)(F)F)C(F)(F)F | Doebelin et al., 2016 DOI:10.1002/cmdc.201600491 | ID:SR19548 | |
| Software, algorithm | HDX-Workbench | Pascal et al., 2012 DOI:10.1007/s13361-012-0419-6 | N/A | |
| Software, algorithm | Prism 5 | GraphPad | N/A | |
| Software, algorithm | R Studio | R studio Team (2015) | N/A | |
| Software, algorithm | PHENIX | Adams et al., 2010 DOI:10.1107/S0907444909052925 | N/A | |
| Software, algorithm | iMosflm | Battye et al., 2011DOI:10.1107/s0907444910048675 | N/A |
Additional files
-
Supplementary file 1
X-ray Co-crystallographic statistics (related to Figures 1 and 2).
Data collection and refinement statistics are shown for SR2211, SR19265, and SR19355 co-crystal structures.
- https://doi.org/10.7554/eLife.47172.012
-
Supplementary file 2
Summary of full time course differential HDX-MS (related to Figures 1 and 2).
Data for control and test conditions were averaged across all timepoints and subtracted to generate differential values. Error was propagated and is shown in parentheses.
- https://doi.org/10.7554/eLife.47172.013
-
Supplementary file 3
The two timepoint HDX-MS screening data were treated and summarized in an. xlsx file (related to Figures 3 and 4).
The DMSO tab shows values for apo protein collected for every compound. The Differential tab shows the Δ%D value collected for each compound. The Differential_SEM tab shows the propagated standard error of the mean (SEM) for each measurement. The ExperimentalData and ExperimentalData_SEM tabs show the biochemical data measurements and SEM for each compound that was used for correlation analysis. The ReducedDifferential and ReducedDifferential_SEM tab shows perturbation values and SEM that are statistically significant from 0 with multiple testing correction FDR > 0.05 and absolute value greater than 5 Δ%D.
- https://doi.org/10.7554/eLife.47172.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47172.015





