Comment on 'Single nucleus sequencing reveals evidence of inter-nucleus recombination in arbuscular mycorrhizal fungi'
Figures
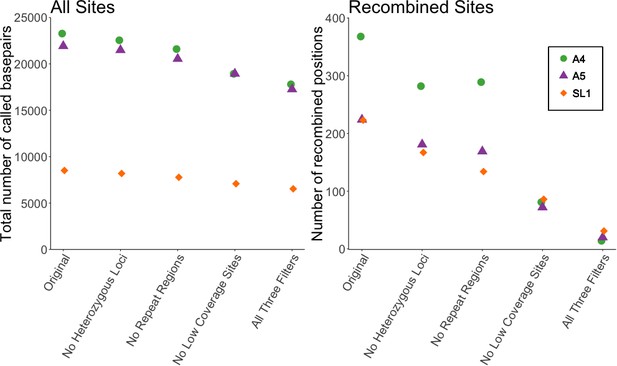
Filtering SNP data shows a decrease of 91% in number of recombined sites, but only ~20% decrease in total sites.
Left panel shows the effect of filtering on all sites included in Supplementary file 6 of Chen et al. (2018a), while right shows the effect on the number of recombined positions. Recombined positions identified based on second criterion in Figure 1—figure supplement 1. Different symbols show the effect on the three different strains (A4, A5, and SL1) used in our re-analysis.
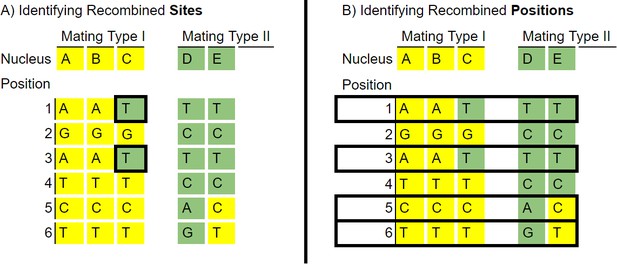
Example of the two criteria used to identify recombination.
To identify individual recombined sites, a parsimony criterion, used for Figure 2B was applied as seen in A), where the recombined nucleus was determined based on the majority genotype for a given mating type. Note that positions 5 and 6 for nucleus E are not identified as there is one nucleus of mating type II with each genotype, this criterion cannot identify sites when there are equal number of nuclei of a mating type with each genotype. To more fully identify recombination positions, we also used a criterion of a difference in genotype between nuclei of the same mating type. This criterion is capable of identify examples such as position 5 and 6, but does not identify which sites are recombined.
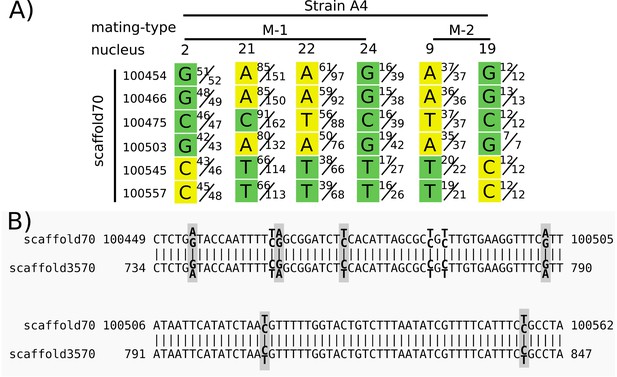
Heterozygous positions in single nuclei from a duplicated region were treated as homozygous in Chen et al. (2018a) and reported as evidence for a long stretch of recombination.
Panel (A) shows the base calls for positions on scaffold 70 for six nuclei (four nuclei of mating type M-1 and two of M-2, as indicated in the top row). Each base call was assigned to a mating type class (green or yellow) in Chen et al. (2018a) based on an unspecified criterion. Variation between nuclei of the same mating type (e.g. variation among nuclei 2, 21, 22, and 24) is interpreted as recombination. We used their Illumina reads to show the ratio of reads supporting alternative nucleotides for each position. For example, in strain 4, mating-type 1, nucleus 2, position 100454, in Chen et al. the base was called as a G with a mating type ‘green’, and 51 of 52 reads matched G. However, for nucleus 21, only 85 of 151 reads supported an A at that position, while the other 66 supported a G. Panel (B) shows the alignment of the region shown in (A) with its best BLAST hit region on scaffold 3570. Heterozygous sites in the mapped reads of the dikaryon are indicated in bold, with the two alternate and reference bases shown slightly above/below. Gray boxes surround those sites included in (A). Note that both regions are heterozygous at the same aligned sites, and with the same alternate base for each heterozygous site. Graphic of (A) modified from Figure 3 of Chen et al. (2018a), with the addition of nuclei 21 and 24.
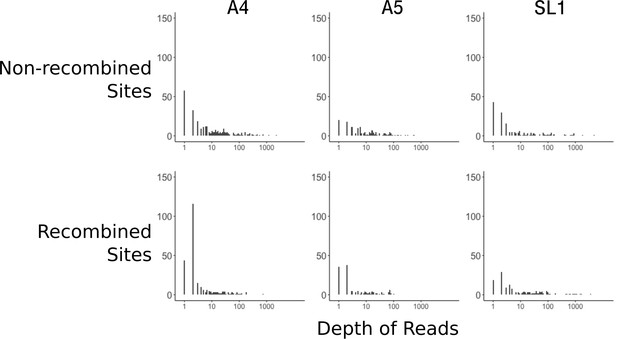
Recombined sites are overrepresented for low coverage sites.
Top row: Distribution of sub-sampled read depths for non-recombined sites of individual nuclei for the three isolates, showing decreased coverage overall for SL1. Bottom row: Distribution of read depths for recombined sites does not mirror the distribution of random sites. Sites identified as recombined based on the parsimony criterion diagrammed in Figure 1—figure supplement 1. Note that read depth is plotted on a log scale.
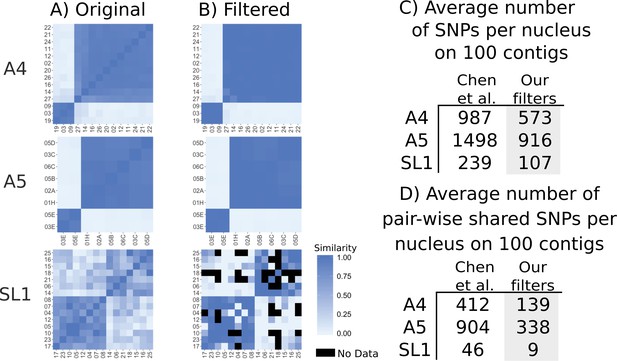
Genetic similarity of SL1 is strongly affected by SNP filtering methods.
(A) left column panels show heatmaps generated from original data presented in Supplementary file 6 of Chen et al. (B) Right panels show data after filtering. Black squares represent pairs that do not share any SNPs. (C) Average number of SNPs in the dataset from Chen et al. (2018a) and after filtering. Note that SL1 has the lowest number of SNPs due to low sequencing breadth. (D) Average number of pairwise overlapping SNPs per nucleus, note that after filtering nuclei from SL1 have fewer than 10 SNPs overlapping on average, again due to low sequencing breadth.
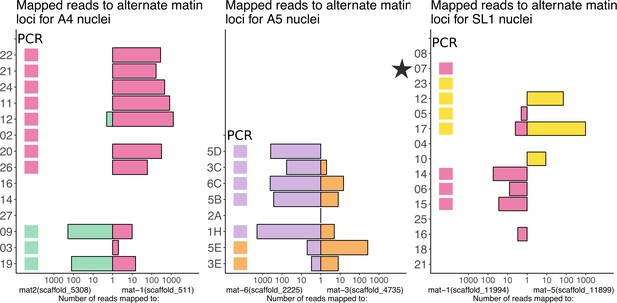
The critical mating type of nucleus 07 (black star) from SL1 is unsupported by short-read data.
The two mating type regions for A4, A5, and SL1 were identified in silico with BLAST searches using the primer sequences and methodology of Ropars et al. (2016). Two mating type loci were identified for each of the three isolates. Reads mapping to either of the two alternate mating type loci are shown below on a log scale. Colors are according to Figure 3 of Chen et al. (2018a), and colored boxes near labels indicate mating type based on PCR amplification as reported in Chen et al. (2018a). Figure 4—figure supplement 1—source data 1. contains a list of all the mating loci identified in A4, A5, and SL1 genome assemblies. This figure shows a general congruence between PCR validated mating types and the number of reads that mapped to either of the two alternate mating loci. For isolate A4, for all nuclei with aligned reads, the PCR results matched the majority of the reads (note the log scale), and 02 and 03 were successful for PCR with no short reads mapped. For isolate A5, all nuclei with a PCR amplified mating type also had a majority of reads mapping to one mating type. Nuclei from SL1 had less success with both short read or PCR, likely due to decreased amplification breadth. Critically, nuclei 07 from SL1 (starred) had no short-reads mapped to either mating locus.
-
Figure 4—figure supplement 1—source data 1
Mating loci identified in A4, A5, and SL1 genome assemblies.
Ths file includes the locations of identified loci, and primer sequences used for BLAST searches. These locations were used for aligning reads from Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.47301.010
Tables
Lengths of recombination events before and after additional filtering.
https://doi.org/10.7554/eLife.47301.004| Original data | Filtered data | |||||
|---|---|---|---|---|---|---|
| Isolate (Mating types) | Number of recombined positions* | Number of recombined blocks (>1 consecutive SNP recombined) | Number of SNPs of longest recombined block (length in bp) | Number of recombined positions* | Number of recombined blocks (>1 consecutive SNP recombined) | Number of SNPs of longest recombined block (length in bp) |
| A4 Mat-1/Mat-2 | 54/314 | 33 | 5 (1131) | 0/14 | 0 | 1 (1) |
| A5 Mat-3/Mat-6 | 41/183 | 18 | 22 (2145) | 2/18 | 2 | 6 (670) |
| SL1 Mat-5/Mat-1 | 111/112 | 20 | 16 (1872) | 22/9 | 4 | 6 (429) |
-
*numbers before/after the slash separate the two mating types, listed in the leftmost column. Number calculated based on the criteria shown in Figure 1—figure supplement 1A. Table 1—source data 1 contains a list of all the recombined blocks identified.
-
Table 1—source data 1
Recombined block locations.
List of all recombined blocks identified, which was used for Table 1.
- https://doi.org/10.7554/eLife.47301.005
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47301.011





