Interspecies interactions induce exploratory motility in Pseudomonas aeruginosa
Figures
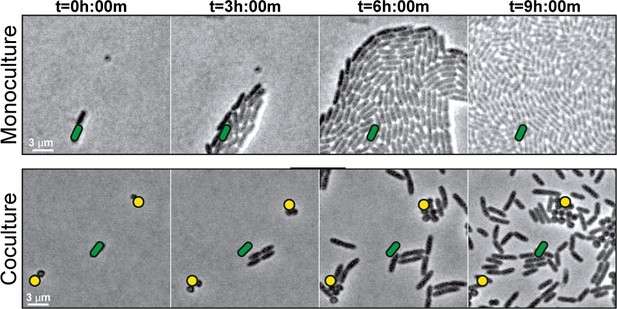
S. aureus increases P. aeruginosa motility.
Live-imaging of polymicrobial interactions. P. aeruginosa (rod-shaped) was inoculated between a coverslip and an agarose pad, either in monoculture (top) or in coculture with equal numbers of S. aureus (cocci-shaped, bottom). Images were acquired every 15 m for 9 hr. Representative snap-shots of Video 1 (top) and Video 2 (bottom) are shown. Founding cells identified in the first frame are indicated with green rods (P. aeruginosa) or yellow circles (S. aureus). The location of the founding cell is indicated in each subsequent frame for positional reference. At t = 9 h:00 m, the founding P. aeruginosa cell has moved outside the field of view.
WT S. aureus in monoculture.
Duration 8 hr. Acquisition interval 15 m. Playback speed 3054x.
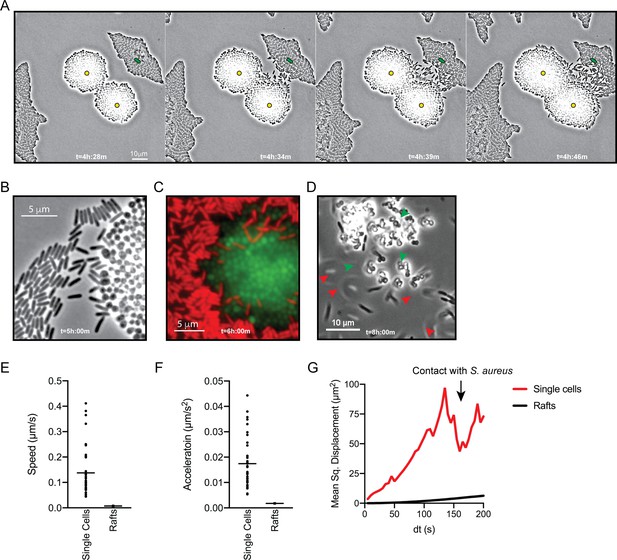
P. aeruginosa adopts an exploratory mode of motility in the presence of S. aureus.
Live-imaging of P. aeruginosa with WT S. aureus (Video 3). (A) Montage of representative snap-shots are shown beginning at 4 hr:28 m. Founding cells identified in the first frame are indicated with green rods (P. aeruginosa) or yellow circles (S. aureus). The location of the founding cell is indicated in each subsequent frame for positional reference. (B) Snap-shot at 5 hr, zoomed in to visualize single-cells. (C) Snap-shot at 6 hr of P. aeruginosa (mKO, red) and S. aureus (GFP, green) illustrating P. aeruginosa invasion into S. aureus colonies. (D) Snap-shot of coculture at 8 hr, showing disruption of S. aureus colonies (green arrows) and swift-moving P. aeruginosa cells out of the plane of focus (red arrows). Single P. aeruginosa cells and the leading edge of rafts were tracked in the presence of S. aureus and the speed (µm/s), acceleration (µm/s2), and mean squared displacement (µm2) for four independent videos are indicated, respectively, in (E – G).
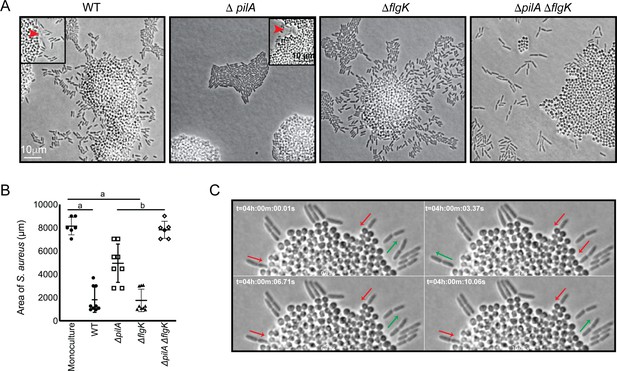
P. aeruginosa exploratory motility is driven by type IV pili.
Live-imaging of P. aeruginosa with WT S. aureus. (A) Representative snap-shots of coculture with WT S. aureus and P. aeruginosa (WT, ΔpilA, ΔflgK, and ΔpilA ΔflgK, left to right, Videos 4 and 5 and Figure 3—videos 1 and 2 respectively) are shown at t = 4.5 hr. Boxed insets show swift-moving P. aeruginosa cells out of the plane of focus (red arrows). (B) The area of S. aureus per frame in monoculture or in the presence of the indicated P. aeruginosa strain was calculated at t = 5 hr by dividing the total area occupied by S. aureus in a single frame by the number of S. aureus colonies. A minimum of four videos were analyzed per condition. The mean and standard deviation are indicated. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between S. aureus in monoculture and in the presence of either WT P. aeruginosa or ΔflgK; b indicates a statistically significant difference between ΔpilA and ΔpilA ΔflgK. (C) Representative snap-shots of Video 4, WT P. aeruginosa and S. aureus beginning at 4 hr with 50 ms intervals, showing back-and-forth motion. Red arrows indicate when a P. aeruginosa cell is moving in towards the S. aureus colony and green arrows indicate when a cell is moving away.
Live-imaging of P. aeruginosa ΔpilA ΔflgK mutant in monoculture.
Representative snapshot at t = 4 hr.
P. aeruginosa ΔflgK in coculture with WT S. aureus.
Duration 30 s. 4.5 hr post inoculation. Acquisition interval 50 ms. Playback speed 3x.
P. aeruginosa ΔpilA ΔflgK in coculture with WT S. aureus.
Duration 30 s. 4.5 hr post inoculation. Acquisition interval 50 ms. Playback speed 3x.
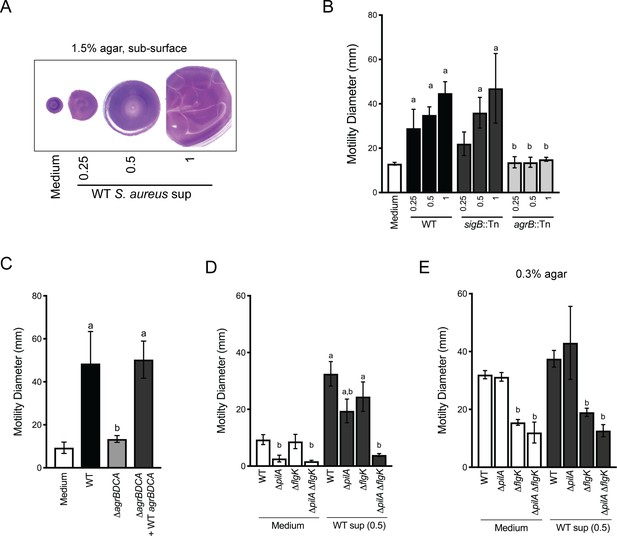
Agr-regulated secreted S. aureus factors increase P. aeruginosa motility.
(A – D) Motility of WT P. aeruginosa was monitored by macroscopic sub-surface inoculation assays in the presence of medium alone or cell-free supernatant from the indicated S. aureus strains. (A) illustrates representative motility zones stained with crystal violet for visualization. The dilution factor of the supernatant is indicated in A and B. Undiluted supernatant was used in C. In D and E, the motility of the indicated P. aeruginosa mutants was analyzed in the presence of medium alone or supernatant derived from WT S. aureus (0.5 dilution) under 1.5% agar (D) or within 0.3% agar (E). The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between the motility observed in the presence of S. aureus supernatant compared to medium alone, and b indicates a statistically significant difference (p≤0.05) between the motility observed in the mutant strain (S. aureus mutants in B and C, P. aeruginosa mutants in D and E) compared to the parental.
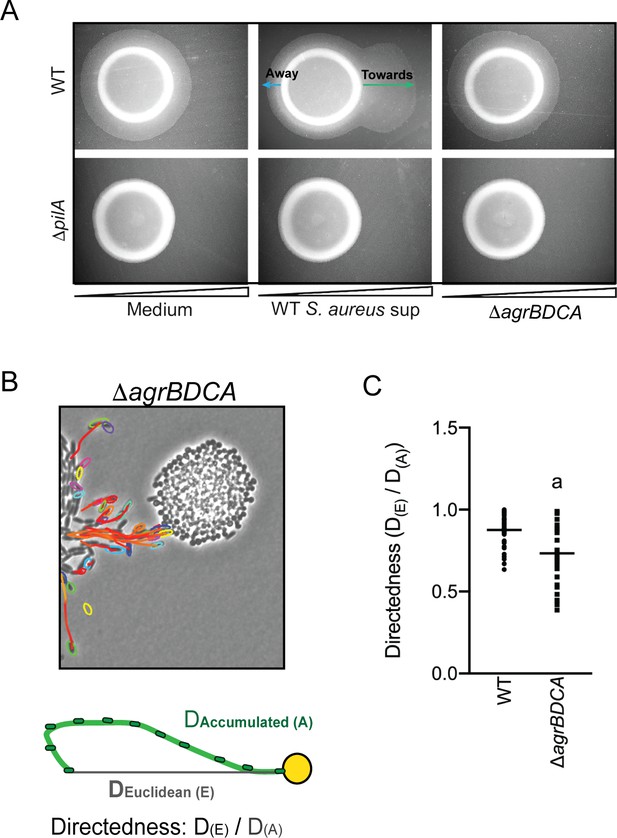
P. aeruginosa biases the directionality of movement up a concentration gradient of Agr-regulated secreted factors.
(A) A concentration gradient of either S. aureus growth medium (TSB), S. aureus supernatant derived from WT or ΔagrBDCA was established by spotting onto the surface of the agar and allowing a concentration gradient to establish by diffusion for approximately 24 hr, prior to spotting P. aeruginosa onto the agar (6 mm to the left) and surface-based motility imaged after 24 hr. Representative images of at least three independent experiments are shown. (B) Example of live-imaging of WT P. aeruginosa with S. aureus ΔagrBDCA with tracks of single-cells shown and schematic illustrating the methods for calculating the directedness. Single P. aeruginosa cells were tracked from first the frame a cell exited the raft to the frame where it first encounters S. aureus. The accumulated track distance, D(A), was measured for at least 30 cells in four independent videos and compared to the Euclidean distance, D(E), between the position of the cell in the first and last frame tracked. The ratio of D(E)/D(A) (Directedness) is shown in (C) for P. aeruginosa moving towards WT S. aureus compared to ΔagrBDCA with the mean indicated. Statistical significance was determined by an unpaired Student’s t-test (a = P ≤ 0.05).
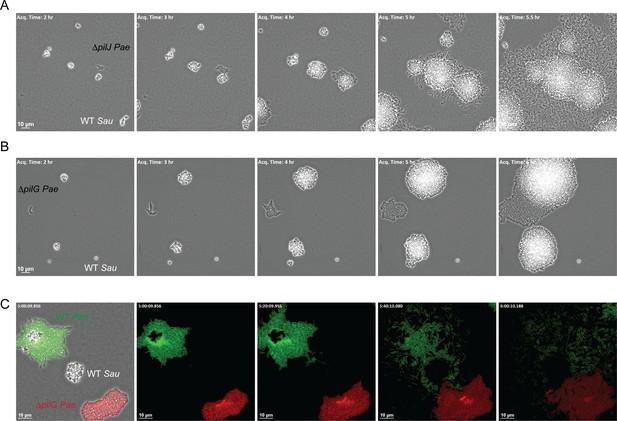
The CheY - like response regulator, PilG modulates P. aeruginosa response to S. aureus.
Representative snap-shots of P. aeruginosa ΔpilJ (A) and ΔpilG (B) in coculture with WT S. aureus. In (C), representative snap-shots of WT P. aeruginosa (GFP, green) with P. aeruginosa ΔpilG (mKate, red) and WT S. aureus (unmarked), visible only in the first frame (left) with phase contrast overlay.
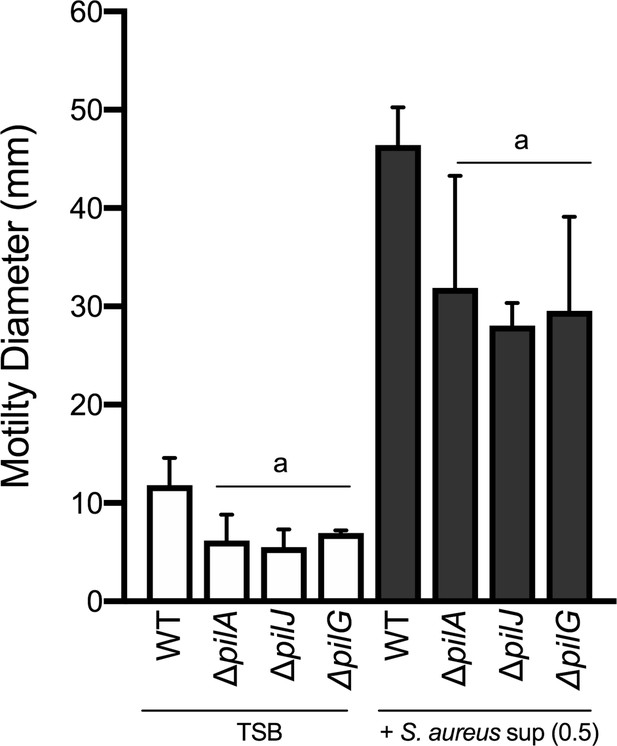
Motility of WT, ΔpilA, ΔpilJ, and ΔpilG P. aeruginosa by macroscopic sub-surface inoculation assays in the presence of medium alone (TSB) or cell-free supernatant from WT S. aureus strains.
The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between the motility observed in the mutant P. aeruginosa strains compared to the parental (WT).
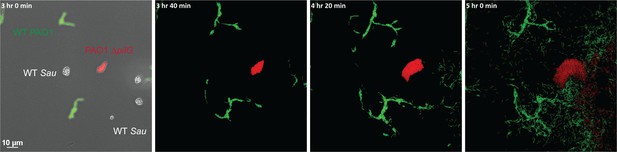
Live-imaging of PAO1 ΔpilG in coculture with WT PAO1 and WT S. aureus.
Representative snap-shots of P. aeruginosa WT PAO1 (GFP, green), PAO1 ΔpilG (mKate, red) in coculture with WT S. aureus (phase contrast only, in first frame, left) showing diminished response to S. aureus in comparison to WT PAO1.
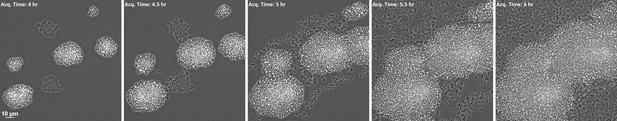
Live-imaging of PA14 ΔpilG in coculture with increased S. aureus inoculum.
Representative snap-shots of P. aeruginosa ΔpilG in coculture with WT S. aureus showing increased S. aureus inoculum promotes motility in a ΔpilG mutant.
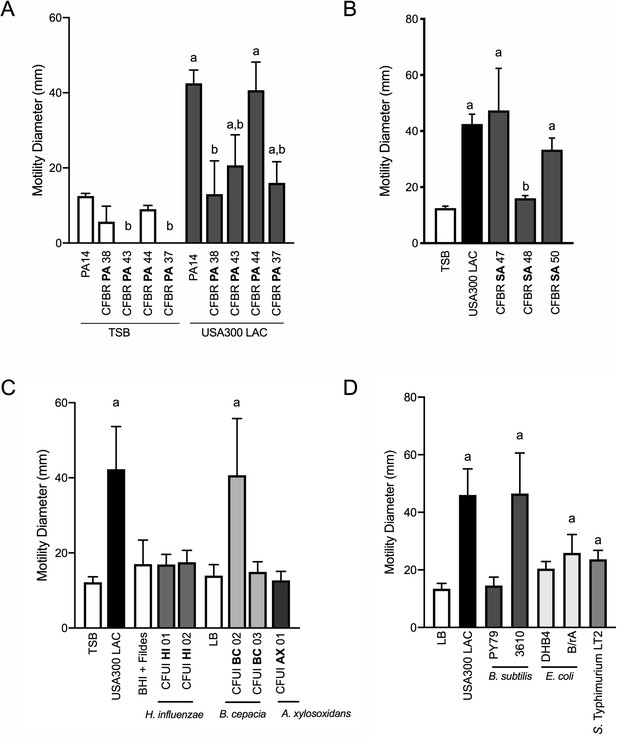
P. aeruginosa modulates motility in response to a range of CF clinical and non-clinical bacterial species.
The motility of clinical P. aeruginosa isolates (CFBR PA 38, 43, 44, and 37), in the presence of cell-free supernatant derived from S. aureus USA300 LAC (0.5 dilution) is indicated, in comparison to P. aeruginosa PA14 (A). The motility of P. aeruginosa laboratory strain PA14 was monitored in the presence of undiluted cell-free supernatant from clinical S. aureus CF isolates, CFBR SA 47, 48, and 50 (B), CF clinical isolates: H. influenzae, B. cepacia, and A. xylosoxidans (C), and non-CF species: E. coli, B. subtilis, and S. Typhimurium (D). Growth medium alone for each species was used as a negative control (TSB: S. aureus, BHI + Fildes: H. influenzae, LB: B. cepacia, A. xylosoxidans, B. subtilis, E. coli, and S. Typhimurium). The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test – a indicates a statistically significant difference (p≤0.05) between the motility observed in the presence of S. aureus supernatant compared to medium alone (A–D), and b indicates a statistically significant difference (p≤0.05) between the motility observed in the CF isolates, in comparison to laboratory strains (P. aeruginosa laboratory strain PA14 in (A) and S. aureus USA300 LAC in (B)).
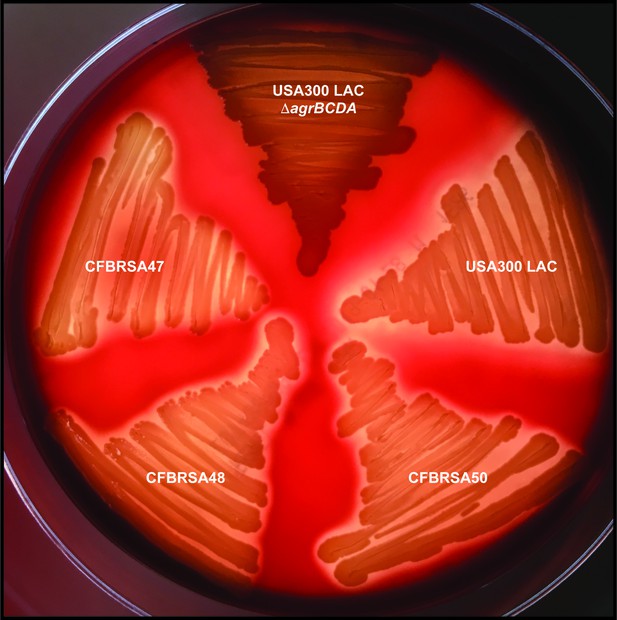
Clinical CF S. aureus isolates retain β-hemolysis.
Laboratory S. aureus isolates USA300 LAC (WT) and ΔagrBDCA and clinical CF S. aureus isolates (CFBRSA47, 48, and 50) were grown on sheep’s blood agar and examined for hemolysis of red blood cells surrounding the colonies.
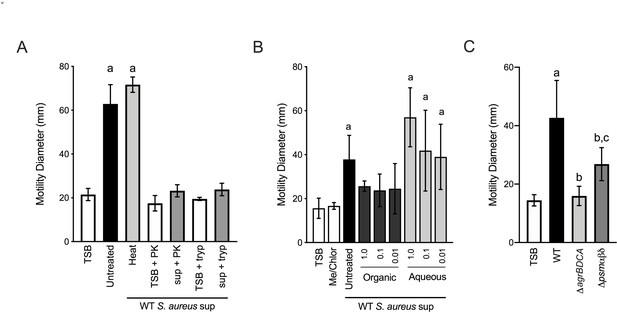
Phenol soluble modulins contribute to S. aureus-induced P. aeruginosa motility.
Motility of WT P. aeruginosa was monitored by macroscopic sub-surface inoculation assays in the presence of medium alone or cell-free supernatant derived from WT S. aureus treated with either heat, proteinase K (PK), or trypsin (tryp) in (A), methanol/chloroform extraction of the supernatant and subsequent analysis of the organic and aqueous fraction (methanol-chloroform (2:1, Me/Chlor) was included as the vehicle control) (B), or in the presence of untreated supernatant derived from WT or the indicated S. aureus mutants (0.5 dilution) in (C). The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between the motility observed in the presence of S. aureus supernatant compared to medium alone, b between the motility observed in the mutant strains compared to the parental, and c between the ΔagrBDCA and Δpsmαβδ mutants.
Videos
WT P. aeruginosa in monoculture.
Duration 8 hr. Acquisition interval 15 m. Playback speed 3054x.
WT P. aeruginosa in coculture with WT S. aureus.
Duration 8 hr. Acquisition interval 15 m. Playback speed 3054x.
WT P. aeruginosa in coculture with WT S. aureus.
Duration 10 m. 4 hr post inoculation. Acquisition interval 5 s. Playback speed 50x.
WT P. aeruginosa in coculture with WT S. aureus.
Duration 30 s. 4.5 hr post inoculation. Acquisition interval 50 ms. Playback speed 3x.
P. aeruginosa ΔpilA in coculture with WT S. aureus.
Duration 30 s. 4.5 hr post inoculation. Acquisition interval 50 ms. Playback speed 3x.
WT P. aeruginosa in coculture with S. aureus ΔagrBDCA.
Duration 10 m. 4 hr post inoculation. Acquisition interval 5 s. Playback speed 50x.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DH5α | Life Technologies | supE44 ΔlacU169 (ϕ80dlacZΔM15) hsdR17 thi-1 relA1 recA1 | |
| Strain, strain background (Escherichia coli) | S17 λpir | Life Technologies | TpR SmR recA, thi, pro, hsdR-M+RP4: 2-Tc:Mu:Km Tn7 λpir | |
| Strain, strain background (Escherichia coli) | B/rA | ATCC | ATCC 12407 | Obtained from David Weiss (Iowa) |
| Strain, strain background (Escherichia coli) | MC4100 | PMID: 11677609 | F− (araD139) Δ(lac)U169, strA, thi | Obtained from David Weiss (Iowa) |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 (WT) | PMID: 7604262 | Non-mucoid prototroph SMC232(O’Toole Strain Collection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 mKO | PMID: 28084994 | attB:: PA1/04/03 -mKO Constitutively produces orange fluorescent protein | Obtained from Carey Nadell (Dartmouth) |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilA | PMID: 20233936 | SMC3782 (O’Toole StrainCollection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔflgK | PMID: 28167523 | SMC5845 (O’Toole StrainCollection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilA ΔflgK | PMID: 28167523 | SMC6595 (O’Toole StrainCollection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilJ | PMID: 18178737 | SMC2992 (O’Toole Strain Collection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilG | SMC4375 (O’Toole StrainCollection) | Obtained from Kyle Cady (Dartmouth) | |
| Strain, strain background (Pseudomonas aeruginosa) | PAO1 ΔpilG | PMID: 20008072 | Obtained from Joanne Engel (UCSF) | |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA37 | PMID: 28325763 | Mucoid cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA38 | PMID: 28325763 | Mucoid cysticfibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA43 | PMID: 28325763 | Mucoid cysticfibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA44 | PMID: 28325763 | Non-mucoid cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC (WT) | PMID: 23404398 | USA300 CA-Methicillin resistant strain LACwithout plasmids | Obtained from Ambrose Cheung (Dartmouth) |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC pCM29 | PMID: 20829608 | sarAP1-sGFP | Obtained from Kenneth Bayles (Nebraska) |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC agrB::TnMar | PMID: 23404398 | Obtained from the Nebraska Transposon Mutant Library | |
| Strain, strain background (Staphylococcus aureus) | USA300 LACsigB::TnMar | PMID: 23404398 | Obtained from the Nebraska Transposon Mutant Library | |
| Strain, strain background (Staphylococcus aureus) | USA300 LACΔagrBDCA | This study | See Materials and methods | |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC ΔagrBDCA + WT agrBDCA | This study | See Materials and methods | |
| Strain, strain background (Staphylococcus aureus) | CFBR SA47 | PMID: 28325763 | Cystic fibrosis isolate | Obtained from the Nebraska Transposon Mutant Library |
| Strain, strain background (Staphylococcus aureus) | CFBR SA48 | PMID: 28325763 | Cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Staphylococcus aureus) | CFBR SA50 | PMID: 28325763 | Cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Haemophilus influenzae) | CFUI HI01 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Haemophilus influenzae) | CFUI HI02 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Burkholderia cepacia) | CFUI BC02 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Burkholderia cepacia) | CFUI BC03 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Achromobacter xylosoxidans) | CFUI AX01 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Bacillus subtilis) | PY79 | PMID: 6093169 | Prototrophic derivative of B. subtilis 168 | Obtained from Craig Ellermeier (Iowa) |
| Strain, strain background (Bacillus subtilis) | 3610 | PMID: 11572999 | NCIB 3610:Marburg ‘wild’ isolate | Obtained from Craig Ellermeier (Iowa) |
| Strain, strain background (Salmonella enterica serovar Typhimurium) | LT2 | PMID: 11677609 | Obtained from Bradley Jones (Iowa) | |
| Strain, strain background (Staphylococcus aureus) | USA300 LACΔpsmαβδ | PMID: 26077761 | psmΔα1–4 Δβ1–2 δATG-ATT | Obtained from Daniel Wozniak (OSU) |
| Recombinant DNA reagent | pMAD | PMID: 15528558 | E. coli-S. aureus shuttle vector. OripE194TS ermC blaC bgaB | |
| Commercial assay or kit | Gibson Assembly Cloning Kit | New England BioLabs | E5510 | |
| Software, algorithm | NIS-Elements AR | Nikon | Version 5.2 |
Oligonucleotides used in this study.
| Name | Sequence | Reference |
|---|---|---|
| agr-a | CGCGGATCCTACATAGCACTGAGTCCAAGG | This study |
| agr-b | GCCGTTAACTGACTTTATTATCTTTTTTACACCACTCTCCTCACTG | This study |
| agr-c | CAGTGAGGAGAGTGGTGTAAAAAAGATAATAAAGTCAGTTAACGGC | This study |
| agr-d | CCGGAATTCCAGTTATTAGCAGGATTTTAGC | This study |
Additional files
-
Source data 1
Data for macroscopic twitching motility assays.
- https://cdn.elifesciences.org/articles/47365/elife-47365-data1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/47365/elife-47365-transrepform-v2.docx





