Membrane interactions of the globular domain and the hypervariable region of KRAS4b define its unique diffusion behavior
Figures
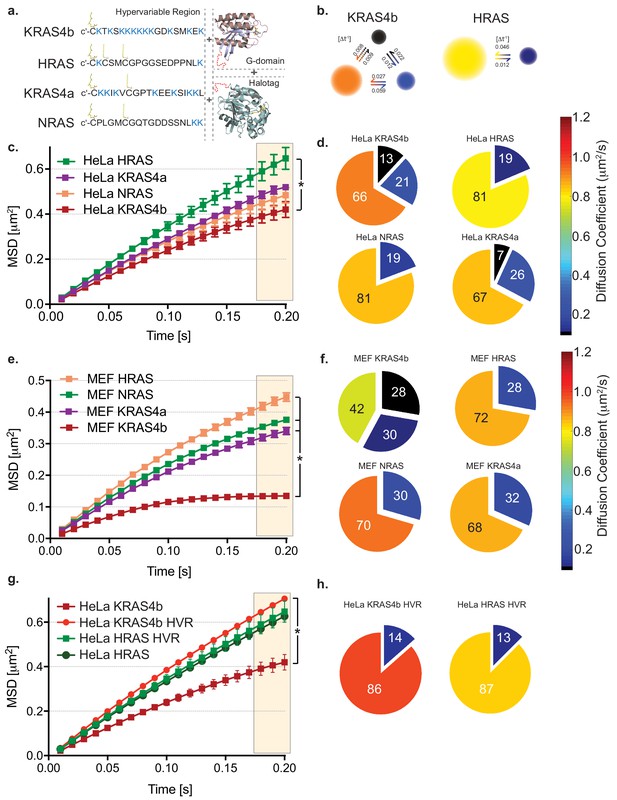
HMM and MSD analysis results of KRAS4b and isoforms in HeLa and MEF cells.
(a) Illustration of combinations of fusion proteins between hypervariable region (HVR) and G-domain of RAS isoforms with HaloTag that were expressed in HeLa and RAS-less MEF cells. (b) Hidden Markov modeling (HMM) using vbSPT of single molecule tracking (SMT) measurements showed different mobile patterns and was described with three and two diffusive states for KRAS4b and HRAS, respectively. Each diffusion (state) coefficient is pseudo-color coded, as marked by the rainbow scale bar and probability of transitions between states per frame rate (Δt = 10 ms). (c) The Mean Squared Displacement (MSD) vs. time plot showed highest confinement of KRAS4b whereas the least for HRAS (* indicates three displacement values under shaded area are significantly different, p<0.05). (d) Diffusion coefficients and occupancy (HMM analysis) from various isoforms of RAS molecules on HeLa cell membrane are summarized in pie charts. (e) MSD analysis on SMT measurements on RAS-less MEF cells for various RAS isoforms (* indicates three displacement values under shaded area are significantly different, p<0.05). (f) The summary of diffusive states from HMM analysis on MEF cells are presented in pie charts for KRAS4b, HRAS, KRAS4a and NRAS. (g) MSD plot showing a significant difference (p<0.05) in confinement (bending of curve) of diffusion between KRAS4b full-length and its truncated HVR, whereas no significant difference was found in case of HRAS. (h) Pie charts show both KRAS4b and HRAS HVRs diffusion were best described by a two-state model using HMM analysis.
-
Figure 1—source data 1
MSD values over time (plotted in Figure 1c) for Halotag-KRAS4b, -KRAS4a, -HRAS, and -NRAS transiently expressed in HeLa cells and recorded with TIRF microscopy.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-data1-v2.txt
-
Figure 1—source data 2
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 1d) of Halotag-KRAS4b, -KRAS4a, -HRAS, and -NRAS transiently expressed in HeLa cells.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-data2-v2.txt
-
Figure 1—source data 3
MSD values over time (plotted in Figure 1e) for Halotag-KRAS4b, -KRAS4a, -HRAS, and -NRAS expressed in isogenic Mouse Embryonic Fibroblast cell pools.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-data3-v2.txt
-
Figure 1—source data 4
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 1f) of Halotag-KRAS4b, -KRAS4a, -HRAS, and -NRAS expressed in isogenic Mouse Embryonic Fibroblasts.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-data4-v2.txt
-
Figure 1—source data 5
MSD values over time (plotted in Figure 1g) for Halotag-KRAS4b, Halotag-KRAS4b HVR (lacking the G domain), Halotag-HRAS, and Halotag-HRAS HVR transiently expressed in HeLa cells.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-data5-v2.txt
-
Figure 1—source data 6
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 1h) of Halotag-KRAS4b HVR and Halotag-HRAS HVR transiently expressed in HeLa cells.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-data6-v2.txt
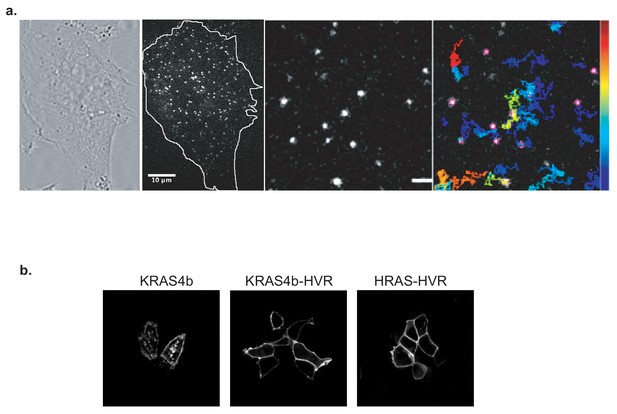
Detection of HaloTagged RAS protein constructs in live cells by single molecule tracking and confocal imaging.
(a) Bright-field and total internal reflection fluorescence (TIRF) microscope image of a HeLa cell expressing HaloTag-KRAS4b after labeling cells with 25pM fluorescent (JF646) HaloTag ligand (Figure 1—video 1). Very sparsely labeled single molecules of membrane-tethered RAS were observed under 100X magnification (scale bar 10 µm). A typical example of 16 µm x16µm region of interest of the plasma membrane in the lamella is shown here that was selected for ultra-fast video imaging. Tracks were obtained from TrackMate software and displayed in pseudo-color according to the residence time of each particle at the membrane (scale bar 2 µm) (Figure 1—video 2). (b) HaloTag-RAS, HVR and its mutant fusion proteins were transiently expressed in HeLa cells to examine and confirm membrane localization using a confocal microscope.
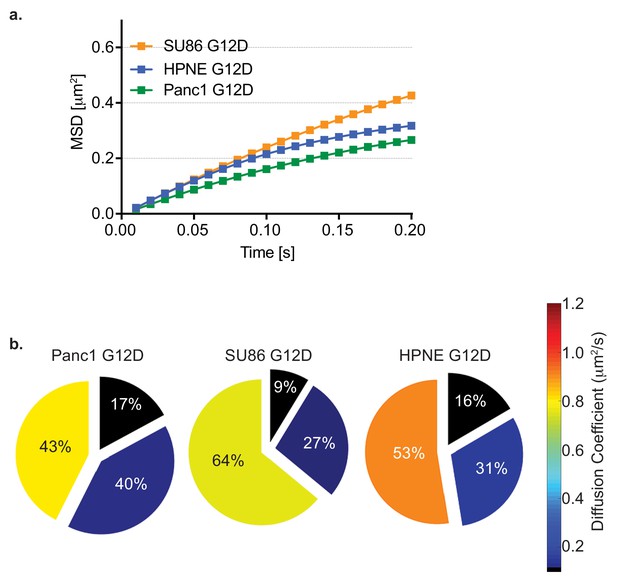
HMM and MSD analysis results of KRAS4b G12D diffusion in cancer cell lines.
HaloTag-KRAS4b G12D diffusion was measured in different pancreatic cancer cell lines (SU.86.86, hTERT-HPNE and PANC-1 harboring mutant KRAS G12D). (a) MSD profile of diffusion of PANC-1 , hTERT-HPNE and SU.86.86 cells. (b) Pie charts indicating diffusion coefficients and occupancy (HMM analysis) of Halo-KRAS4b in background of mutant KRAS4b in pancreatic cancer cell lines.
-
Figure 1—figure supplement 2—source data 1
MSD values over time (plotted in Figure 1—figure supplement 2a) for overexpressed, exogenous Halotag-KRAS4b G12D in a panel of pancreatic cancer cell lines with existing KRAS4b G12D mutations (SU.86.86, hTERT-HPNE , and PANC-1).
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-figsupp2-data1-v2.txt
-
Figure 1—figure supplement 2—source data 2
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 1—figure supplement 2b) for overexpressed, exogenous Halotag-KRAS4b G12D in a panel of pancreatic cancer cell lines with existing KRAS4b G12D mutations (SU.86.86, hTERT-HPNE , and Panc-1).
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-figsupp2-data2-v2.txt
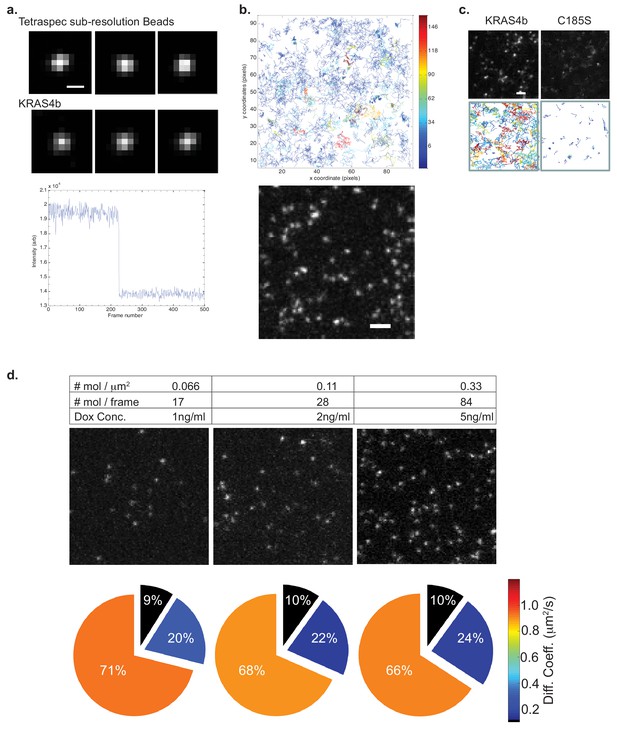
Single detection, trajectories of KRAS4b diffusion and Dox-induced KRAS4b diffusion.
(a) Single-molecule detection was confirmed by comparing the point spread function (PSF) of a single sub-resolution bead vs. RAS molecule under our TIRF microscope (top panel), and by single-step photobleaching of fluorescent RAS particles, as depicted in the profile. (b) Example of a typical membrane area (16 µm x 16 µm) showing HaloTag-KRAS4b tracks obtained during ultrafast video imaging for 10 s. (c) C185S mutant, which is devoid of farnesylation modification at the C-terminus of KRAS4b, could not associate to the plasma membrane as very few particles were observed during TIRF video microscopy (Figure 1—video 3). Note: scale bars in a) represent 500 nm, in b) and c) represent 2 µm. (d) KRAS4b surface density of Dox induced KRAS4b expressed at different levels in HeLa cells; HMM analysis showed no significant difference in its diffusion rates and occupancy at different express levels for KRAS4b.
-
Figure 1—figure supplement 3—source data 1
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 1—figure supplement 3d) of Halotag-KRAS4b for increasing concentrations of doxycycline in a dox-inducible Halotag-KRAS4b HeLa cell pool.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig1-figsupp3-data1-v2.txt
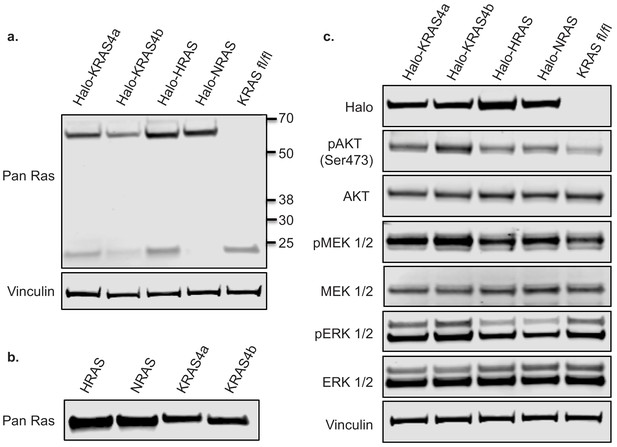
Basal signaling profiles and Ras expression levels of isogenic MEF pools.
(a) Basal RAS expression levels of isogenic MEF pools were analyzed by western blot and compared to the parental MEF cell line, which expresses endogenous murine floxed Kras alone (b) 10 ng purified human RAS protein was analyzed by western blot using a mouse monoclonal pan RAS antibody (c) Basal signaling profiles of isogenic MEF pools were analyzed by western blot alongside the parental line.
TIRF video microscopy of HaloTag-KRAS4b in a live HeLa cell.
16µm x16µm region of interest of the plasma membrane in Hela cell and tracks.
TIRF video microscopy of HaloTag-KRAS4b-C185S mutant in live Hela cell.
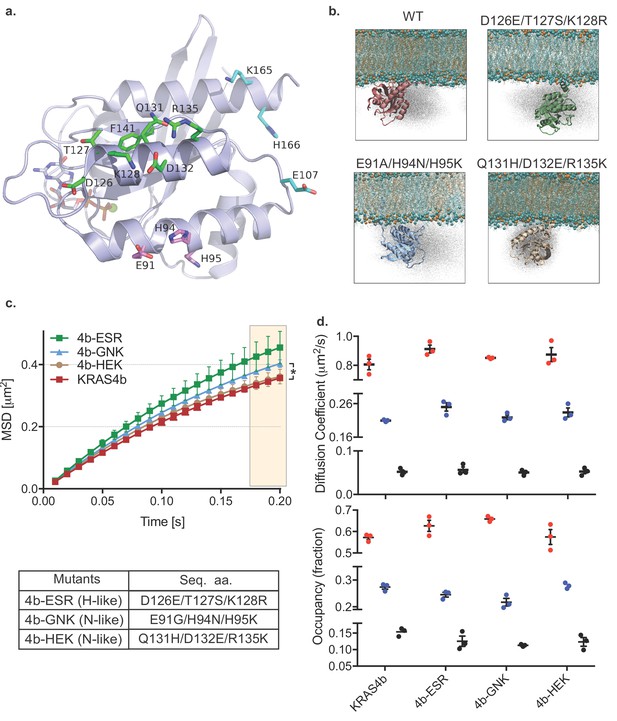
HMM and MSD analysis results of KRAS4b with mutated contact points of G-domain with the lipids and the MD simulation of KRAS4b and the same mutant.
(a) Ribbon diagram of the G-domain of KRAS depicts locations of transitory contact points with the lipids and the following table shows mutations made to the G-domain to mimic H-like or N-like RAS at those key residues. (b) Snapshots from simulations of KRAS4b on lipid membrane (POPC: POPS, 80:20) are shown where G-domain makes transitory contacts with the lipids via salt bridges, with the ghost/shadow representation exhibiting space that G-domain has sampled. (c) MSD plot from the H-like or N-like mutants are relatively less confined compared to WT-KRAS4b (the asterisk * indicates three displacement values under shaded area are significantly different, p<0.05). (d) Diffusion coefficients and occupancy obtained from HMM analysis are compared for each mutant; no significant difference was found between mutants.
-
Figure 2—source data 1
MSD values over time (plotted in Figure 2c) of Halotag-KRAS4b and G-domain mutants 4b-ESR (HRAS-like), 4b-GNK (NRAS-like), and 4b-HEK (NRAS-like).
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig2-data1-v2.txt
-
Figure 2—source data 2
Diffusion coefficients and occupancy fractions obtained by HMM analysis of Halotag-KRAS4b and G-domain mutants 4b-ESR (HRAS-like), 4b-GNK (NRAS-like), and 4b-HEK (NRAS-like).
The three diffusion states (fast, medium, and slow) of each protein were plotted (Figure 2d) by diffusion coefficient (D1, D2, D3), and occupancy fraction (F1, F2, F3).
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig2-data2-v2.txt
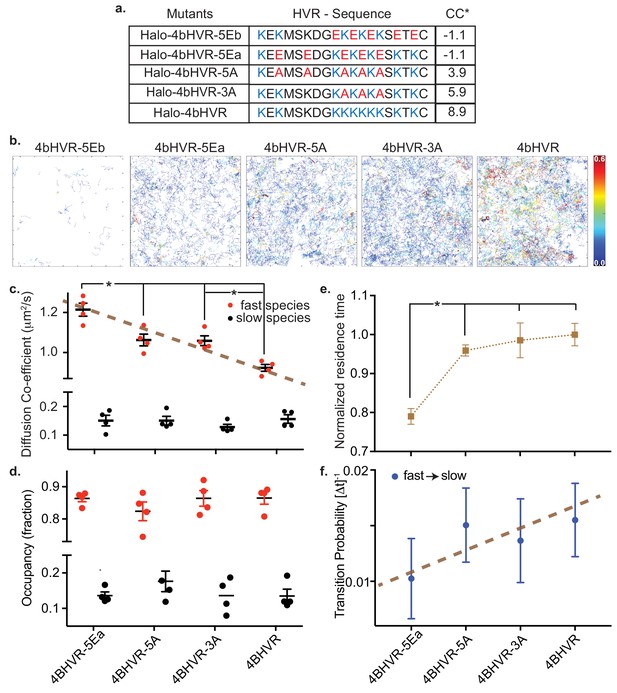
HMM analysis of KRAS4b HVR and mutations that adjusted the charges of the HVR.
(a) Table shows HaloTag HVR (of KRAS4b) constructs over-expressed in HeLa cells by transient transfection with charge-neutral and charge-reversed substitution mutations of lysine residues with varying net charge content (*CC). (b) Single molecule tracks are shown for WT HVR and each of the mutant HVRs, color-coded according to their residence time at the membrane. The color bar encodes time from 0.0 s to 0.6 s. (c and d) Diffusion coefficient and occupancy of the fast and slow diffusing species are plotted in red and black solid circles respectively for each charge-altered mutant. While the fast diffusing species exhibit a gradual, significant (p<0.05) increase in diffusion coefficient as the lysine charges are neutralized and then reversed, the slow species remain the same and the relative fraction of fast and slow diffusing species remain unchanged. (e) Normalized average residence time from more than 5000 tracks for each mutant is shown in the plot. Significant reduction (p<0.05) in residence time indicates impaired association between charge-reversed HVR and membrane. (f) Graph shows transition probabilities from fast to slow diffusive state for each charge-altered mutant.
-
Figure 3—source data 1
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 3c and d) of Halotag-KRAS4b HVR and the charge reversal mutants 5Ea, 5A, and 3A transiently overexpressed in HeLa cells.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig3-data1-v2.txt
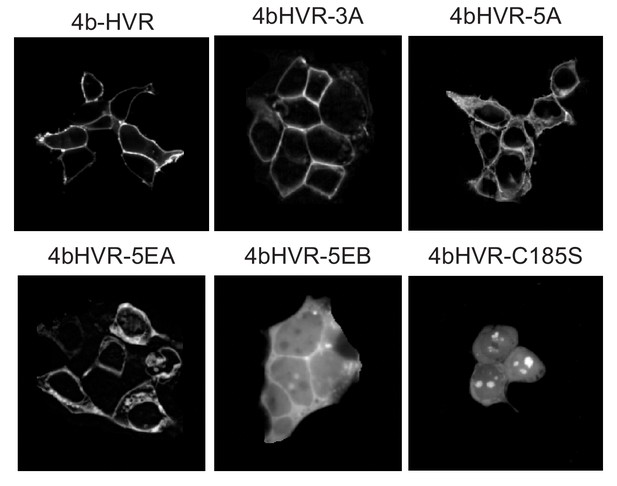
Localization of KRAS4b HVR mutant constructs.
HaloTag-fusion proteins linked to the KRAS4b HVR region were expressed in HeLa cells by transient transfection and visualized by confocal microscopy. Introducing charge deficient mutations in the polylysine region of the KRAS4b HVR reduces membrane localization, most notably in the 5Eb mutant.
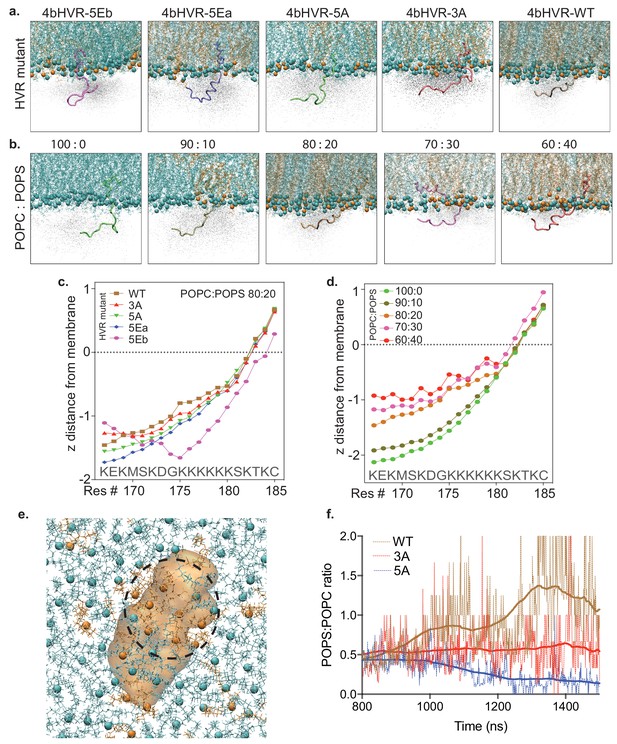
Atomistic simulation of KRAS4b HVR and its mutants interacting with the artificial membrane.
(a) Atomistic simulations from various KRAS4b’s HVR mutants on membrane (POPC: POPS, 80:20) are shown in solid representation where the snapshot most closely represents the average positioning data. In addition, the space HVR has sampled are shown in ghost/shadow representation. (b) Systematic change of POPS from 0% to 40% in the membrane shows increasing proximity of the HVR to the membrane. (c and d) Line plot shows quantification of ‘z’ distance between lipid head groups and individual amino acids (carbon alpha) in the HVR obtained from the simulations. (e) A snapshot from the simulation showing clustering of POPS around the HVR because of electrostatic interaction between positively charged lysine residues and anionic lipid head groups. Blue spheres represent POPC, brown spheres represent POPS, and the 4bHVR is modeled in brown (POPC (80%) – POPS (20%)). (f) Quantification of POPS: POPC ratio in the proximity of the HVR during the time course of simulation. It shows 4bHVR’s ability to concentrate POPS, compared to the charge-deficient mutants, 4bHVR-3A and 4bHVR-5A.
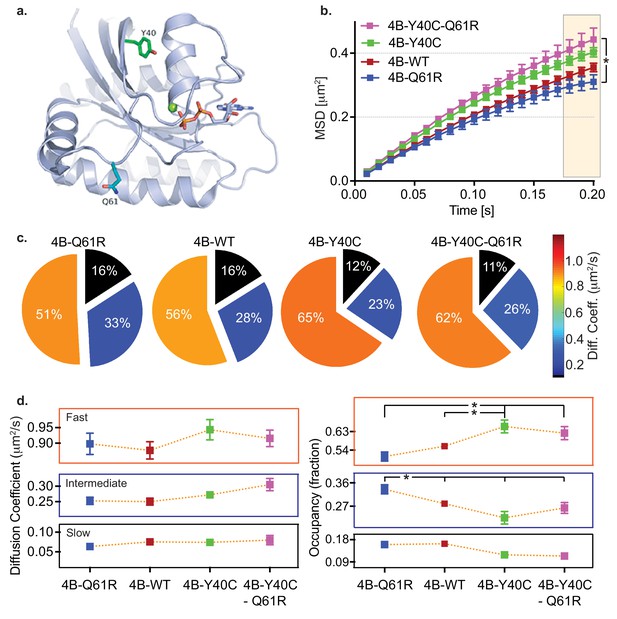
HMM and MSD analysis results of KRAS4b and the oncogenic and RAF binding disabled mutants.
(a) Structural illustration of KRAS4b in ribbon diagram depicts amino acid locations for the mutations Y40C (green), Q61R (blue), and GTP (yellow) (b) MSD plot showed significant difference in diffusion properties between various KRAS4b mutants transiently expressed in HeLa cells. Increased linearity in MSD profile for RAF binding-deficient mutants Y40C and Q61R/Y40C suggests that the effector interaction is another major contributor in confinement of diffusion of KRAS4b. The three displacement values under shaded area between Q61R and Q61R-Y40C mutants are significantly different, p<0.05). (c and d) Pie charts and line plots show the Y40C and Q61R/Y40C mutants have higher occupancy in the fast-diffusing state while lower occupancy the intermediate-diffusing state as determined by vbSPT HMM analysis. Occupancy of fast and intermediate diffusing species is significantly (p<0.05) different between Q61R/WT and Y40C/Q61R-Y40C variants.
-
Figure 5—source data 1
MSD values over time, plotted in Figure 5b, of Halotag-KRAS4b, oncogenic KRAS4b-Q61R, Raf-binding deficient mutant KRAS4b Y40C, and the combination mutant KRAS4b-Y40C-Q61R transiently expressed in HeLa cells.
Mean, standard deviation, and replicate numbers are reported for each protein.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig5-data1-v2.txt
-
Figure 5—source data 2
Diffusion coefficients and occupancy fractions obtained by HMM analysis (plotted in Figure 5c) for Halotag-KRAS4b, -KRAS4b Q61R, -KRAS4b Y40C, and -KRAS4b Y40C-Q61R.
Mean, standard deviation, and replicate numbers are reported for each protein.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig5-data2-v2.txt
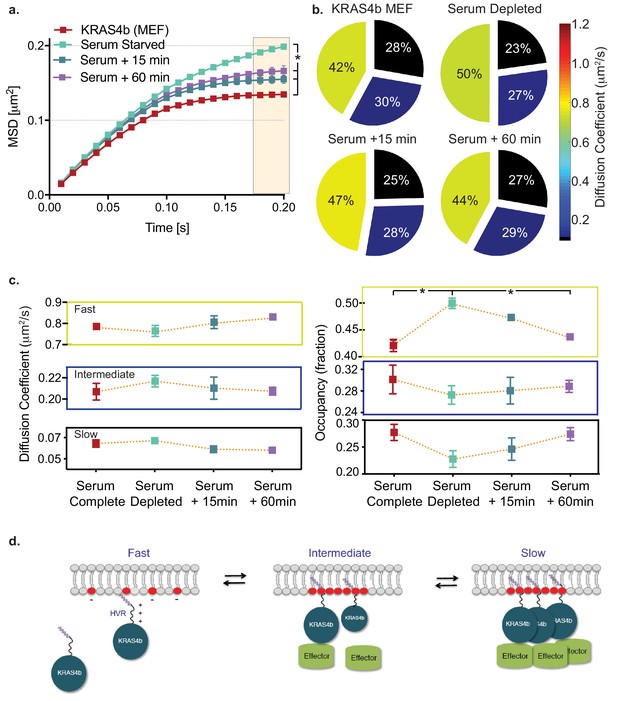
HMM and MSD analysis results of KRAS4b diffusion in MEF in starved and serum complete media rescued conditions.
(a) MSD plot shows relative changes of confinement in diffusion profile of KRAS4b in MEF cells upon serum deprivation (incubated in 0.1% FBS media for 18 hr). MEF cells that are expressing only the KRAS4b isoform, were deprived from serum for 18 hr and then, rescued with 10% FBS containing DMEM media for 15 mins and 60 mins respectively on each coverslip. The three displacement values under shaded area between serum substituted and depleted are significantly different (p<0.05), as indicated by the asterisk ‘*’. (b) vbSPT HMM analysis from same diffusion tracks is displayed on the right-side panel, showing reduced slow-diffusing fraction in serum-deprived state. (c) Line plots of KRAS4b diffusion coefficients and state occupancy in complete media, depleted media and rescued with complete media after 15 and 60 min as determined by vbSPT HMM analysis. Occupancy of fast diffusing species is significantly (p<0.05) different between serum substituted and depleted conditions. (d) Cartoon model showing KRAS4b dynamics in the membrane.
-
Figure 6—source data 1
MSD values over time (plotted in Figure 6a) of Halotag-KRAS4b in complete serum conditions (10% FBS), serum starved (0.1% FBS 18 hr), and after 15 or 60 mins of rescue with 10% FBS serum.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig6-data1-v2.txt
-
Figure 6—source data 2
Diffusion coefficients and occupancy fractions obtained by HMM analysis (visualized in Figure 6b and Figure 6c) for Halotag-KRAS4b in complete serum conditions (10% FBS), serum starved (0.1% FBS 18 hr), and after 15 or 60 min of rescue with 10% FBS serum.
- https://cdn.elifesciences.org/articles/47654/elife-47654-fig6-data2-v2.txt
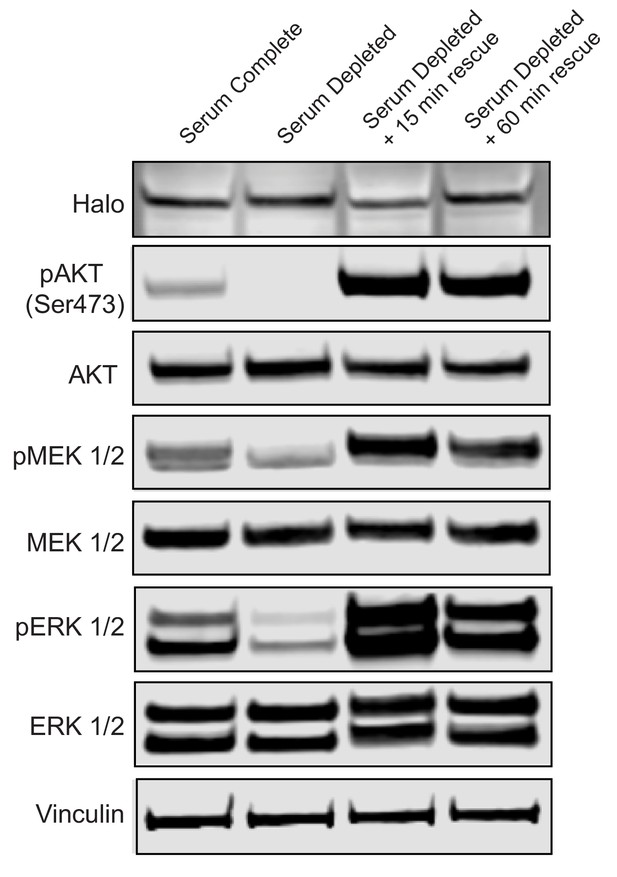
Western blot analysis of KRAS4b signaling under serum starvation and recovery with serum complete media.
Activation of RAS’s downstream signaling (pAKT, pMEK, and pERK) was analyzed by western blot in parallel to single molecule tracking experiments shown in Figure 6. The isogenic MEF pool expressing Halotag-KRAS4b was starved of serum for 18 hr in 0.1% FBS media and rescued with 10% FBS media for a duration of 15 and then 60 min. Serum complete cells remained in 10% FBS media for the duration of the experiment.
Tables
Diffusion rates and percent occupancy in RAS isoforms, and their HVRs.
| Dataset HeLa | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| KRAS4b | 0.95 ± 0.03 | 0.24 ± 0.03 | 0.06 ± 0.01 | 64 ± 2 | 22 ± 2 | 14 ± 3 |
| KRAS4a | 0.82 ± 0.04 | 0.26 ± 0.01 | 0.05 ± 0.01 | 67 ± 2 | 26 ± 3 | 7 ± 2 |
| NRAS | 0.84 ± 0.04 | 0.23 ± 0.08 | 81 ± 2 | 19 ± 1 | ||
| HRAS | 0.81 ± 0.02 | 0.1 ± 0.03 | 81 ± 1 | 19 ± 1 | ||
| KRAS4b HVR | 0.97 ± 0.03 | 0.1 ± 0.05 | 87 ± 3 | 13 ± 3 | ||
| HRAS HVR | 0.82 ± 0.06 | 0.1 ± 0.03 | 87 ± 2 | 13 ± 2 | ||
| NRAS HVR | 0.95 ± 0.08 | 0.15 ± 0.05 | 86 ± 1 | 14 ± 1 | ||
Diffusion rates and percent occupancy of KRAS4b in cancer cell lines
| Dataset KRAS4b | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| HeLa (KRAS4b WT) | 0.95 ± 0.03 | 0.24 ± 0.03 | 0.06 ± 0.01 | 64 ± 2 | 22 ± 2 | 14 ± 3 |
| MEF (KRAS4b WT) | 0.73 ± 0.12 | 0.25 ± 0.09 | 0.05 ± 0.01 | 42 ± 9 | 30 ± 8 | 28 ± 2 |
| PANC-1 (KRAS4b G12D) | 0.84 ± 0.06 | 0.22 ± 0.01 | 0.04 ± 0.01 | 43 ± 6 | 40 ± 4 | 17 ± 2 |
| SU.86.86 (KRAS4b G12D) | 0.8 ± 0.16 | 0.18 ± 0.09 | 0.02 ± 0.01 | 66 ± 14 | 28 ± 9 | 6 ± 5 |
| hTERT-HPNE (KRAS4b G12D) | 0.92 ± 0.06 | 0.25 ± 0.02 | 0.06 ± 0.01 | 55 ± 3 | 29 ± 1 | 16 ± 3 |
Diffusion rates and percent occupancy in full length KRAS4b with increasing concentrations of doxycycline in a dox-inducible HeLa cell pool
| Dataset HeLa | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| DOX 1 ng/mL | 0.96 ± 0.04 | 0.33 ± 0.04 | 0.09 ± 0.01 | 71 ± 1 | 20 ± 2 | 9 ± 1 |
| DOX 2 ng/mL | 0.93 ± 0.02 | 0.26 ± 0.01 | 0.07 ± 0.002 | 68 ± 2 | 22 ± 2 | 10 ± 1 |
| DOX 5 ng/mL | 0.95 ± 0.04 | 0.27 ± 0.01 | 0.07 ± 0.01 | 66 ± 3 | 24 ± 1 | 10 ± 2 |
Diffusion rates and percent occupancy in full length KRAS4b wildtype and with G-domain mutations ESR, GNK, and HEK
| Dataset HeLa | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| KRAS4b | 0.84 ± 0.02 | 0.22 ± 0.01 | 0.05 ± 0.003 | 57 ± 1 | 27 ± 1 | 14 ± 1 |
| KRAS4b ESR (D126E/T127S/K128R) | 0.91 ± 0.04 | 0.25 ± 0.02 | 0.06 ± 0.01 | 63 ± 4 | 25 ± 2 | 12 ± 2 |
| KRAS4b GNK (E91G/H94N/H95K) | 0.85 ± 0.01 | 0.22 ± 0.01 | 0.05 ± 0.01 | 66 ± 1 | 22 ± 2 | 12 ± 1 |
| KRAS4b HEK (Q131H/D132E/R135K) | 0.90 ± 0.06 | 0.24 ± 0.01 | 0.06 ± 0.003 | 58 ± 5 | 28 ± 1 | 14 ± 4 |
Diffusion rates and percent occupancy in full length KRAS4b, the HVR, and HVR mutants in HeLa cells
| Dataset HeLa | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| KRAS4b | 0.95 ± 0.06 | 0.24 ± 0.03 | 0.06 ± 0.01 | 66 ± 2 | 23 ± 2 | 14 ± 3 |
| 4bHVR | 0.98 ± 0.03 | 0.17 ± 0.03 | 87 ± 5 | 13 ± 5 | ||
| 4bHVR-3A | 1.08 ± 0.06 | 0.13 ± 0.04 | 87 ± 6 | 13 ± 5 | ||
| 4bHVR-5A | 1.07 ± 0.04 | 0.15 ± 0.02 | 82 ± 8 | 18 ± 6 | ||
| 4bHVR-5Ea | 1.21 ± 0.05 | 0.15 ± 0.05 | 87 ± 3 | 13 ± 4 | ||
| 4bHVR-5Eb | N/A | N/A | N/A | N/A | N/A | N/A |
Diffusion rates and percent occupancy in full length KRAS4b, KRAS4b Q61R, KRAS4b Y40C, and KRAS4b Y40C-Q61R in HeLa cells
| Dataset HeLa | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| KRAS4b Q61R | 0.90 ± 0.07 | 0.25 ± 0.02 | 0.06 ± 0.01 | 51 ± 5 | 33 ± 3 | 16 ± 1 |
| KRAS4b | 0.88 ± 0.06 | 0.25 ± 0.02 | 0.08 ± 0.01 | 56 ± 2 | 28 ± 8 | 16 ± 1 |
| KRAS4b Y40C | 0.94 ± 0.07 | 0.27 ± 0.02 | 0.07 ± 0.01 | 66 ± 6 | 23 ± 5 | 12 ± 2 |
| KRAS4b Y40C-Q61R | 0.92 ± 0.05 | 0.31 ± 0.04 | 0.08 ± 0.02 | 62 ± 6 | 26 ± 4 | 11 ± 2 |
Diffusion rates and percent occupancy in Ras isoforms, and MEF serum starved cell recovery with serum complete media
| Dataset KRAS4b | Diffusion coefficient (µm2/s) | Occupancy (%) | ||||
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | F1 | F2 | F3 | |
| MEF | 0.73 ± 0.12 | 0.25 ± 0.09 | 0.05 ± 0.01 | 42 ± 3 | 30 ± 3 | 28 ± 4 |
| MEF (srm 0’) | 0.72 ± 0.02 | 0.22 ± 0.02 | 0.07 ± 0.01 | 50 ± 3 | 27 ± 0.8 | 23 ± 3 |
| MEF (srm 15’) | 0.77 ± 0.04 | 0.22 ± 0.02 | 0.06 ± 0.01 | 47 ± 3 | 28 ± 3 | 25 ± 2 |
| MEF (srm 60’) | 0.85 ± 0.03 | 0.21 ± 0.02 | 0.06 ± 0.01 | 44 ± 4 | 29 ± 3 | 27 ± 2 |
Additional files
-
Supplementary file 1
Statistical analysis of data.
For mean-squared displacement plots, an unpaired, two-tailed t-test was performed on three displacement values associated with the later time points in the plots. p-Values are presented to indicate significance in the difference between each pair of conditions. For HMM analysis, diffusion co-efficient and occupancy values are compared using an unpaired, two-tailed t-test to find significance in the difference between pairs of interest. p-Values are presented only for those tests that have passed the significance test (p<0.05).
- https://cdn.elifesciences.org/articles/47654/elife-47654-supp1-v2.txt
-
Supplementary file 2
Table of key resources used in the study.
- https://cdn.elifesciences.org/articles/47654/elife-47654-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/47654/elife-47654-transrepform-v2.docx





