Epigenetic silencing of a multifunctional plant stress regulator
Figures
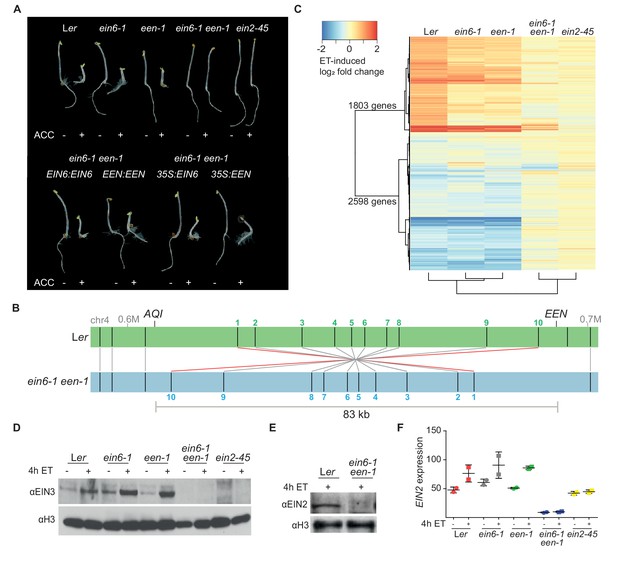
Mutations in two different genes are responsible for the ET-insensitivity in ein6-1 plants.
(A) Triple response phenotype of 3-day-old etiolated seedlings of Ler, ein6-1, een-1, ein6-1 een-1, ein2-45 (upper row) and of the indicated complementation lines, either driven by the respective native promoter or by the Cauliflower mosaic virus 35S promoter (35S) (lower row). Seedlings were grown on control LS media or LS media supplemented with 10 µM ACC. (B) Schematic illustration of a Bionano Genomics Irys optical map of the een-1 inversion region at the end of the fourth chromosome in ein6-1 een-1 (blue) aligned to an optical map of Ler (green). Original output is shown in Figure 1—figure supplement 1D. Nick sites are indicated as black lines within the respective optical map. Matching nick sites between the maps are indicated as gray lines. Numbered nick sites are used to better visualize the inversion event. The approximate position of EEN and AQI is indicated as well. (C) Heatmap visualizes the log2 fold change of expression in Ler, ein6-1, een-1, ein6-1 een-1 and ein2-45 seedlings in response to 4 hr of ethylene (ET) treatment detected by RNA-seq. Differentially expressed genes (DE genes) that are significantly induced (1803 genes) or repressed (2598 genes) after 4 hr of ET treatment in Ler seedlings are shown. Cluster dendogram below the heatmap indicate similarities between the tested genotypes. (D), (E), Western blot analysis of nuclear extracts of 3-day-old etiolated Ler, ein6-1, een-1, ein6-1 een-1 and ein2-45 seedlings that were either treated for 4 hr with hydrocarbon-free air (control) or ET gas. Antibodies against EIN3 and EIN2 were used to detect nuclear-localized EIN3 (D) and EIN2C (E), respectively. Amounts of histone H3 were detected with an anti-H3 antibody and served as a loading control. (F) EIN2 expression in response to 4 hr of ET treatment in the indicated genotypes using RNA-seq is shown. EIN2 transcripts can still be detected in ein2-45 mutants because they only harbor a point mutation in the C-terminus of EIN2 (Beaudoin et al., 2000). Expression is measured in TPM (Transcripts Per Kilobase Million) and results from two biological replicates are shown.
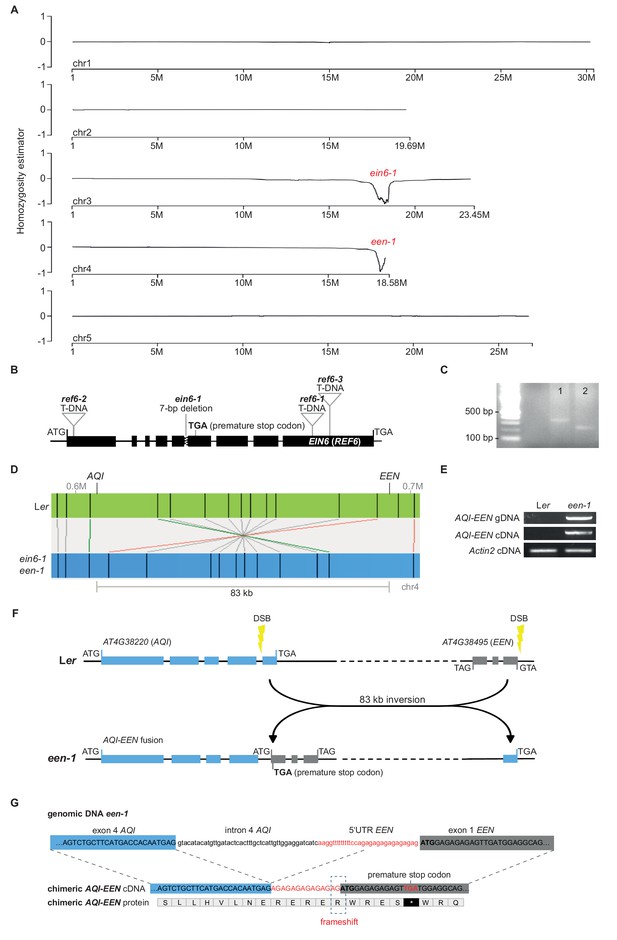
Characterization of the two causal mutations in ein6-1 een-1 double mutants.
(A) SHOREmap analysis of the ET-insensitivity quantitative trait locus (QTL) revealed the genomic location of the ein6-1 and een-1 mutation. At even allele frequency of both parents (Col-0, Ler) the homozygosity estimator is 0. At +1 it’s homozygous for Col-0; at −1 is homozygous for Ler. The five chromosomes of the Arabidopsis genome are shown in 5 Mb increments. (B) Graphic illustration shows the known T-DNA insertions (ref6-1, ref6-2, ref6-3) and the 7bp-deletion (ein6-1) in the EIN6 (REF6) gene. The premature stop codon in the fifth exon of EIN6 (REF6) that results from the 7bp-deletion is indicated. (C) Agarose gel image shows the TAIL-PCR amplicons of the secondary PCR (1) and tertiary PCR (2) amplified with the AD2 primer. 2 kb DNA ladder indicates the approximate size of the amplicons. (D) Optical map of ein6-1 een-1 aligned to Ler indicates the 83 kb inversion event on the fourth chromosome. (E) Agarose gel image shows the chimeric AQI-EEN fusion that is only present in een-1 mutants, on the gene (upper row) and cDNA (middle row) level detected by PCR and RT-PCR, respectively. Level of Actin2 cDNA (lower row) indicates equal loading of Ler and een-1 cDNA. (F) Scheme of the inversion event that led to the AQI-EEN fusion is illustrated. The fast neutron mutagenesis induced a double strand break (DSB) in the start of fourth intron of AQI and another DSB in the 5’UTR of EEN. Consequently, incorrect repair of the DSBs resulted in a 83 kb inversion event leading to the fusion of AQI with EEN. (G) Detailed illustration of the AQI-EEN fusion at the gene, cDNA and protein level in nucleotide and amino acid resolution, respectively. Sequence comparison between the AQI-EEN fusion at gene and cDNA level revealed that the 5’UTR of EEN was incompletely spliced after the fusion with the residual fourth intron of AQI. The remaining 14 bp of the 5’UTR became part of the coding sequence causing an open reading frame shift in the first exon of EEN.
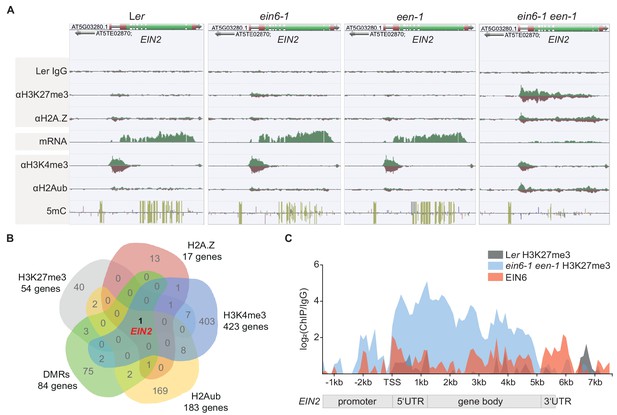
A repressive chromatin environment at EIN2 down-regulates its expression.
(A) Genome browser screenshot visualizes the levels of the depicted chromatin features at the EIN2 gene in untreated 3-day-old etiolated Ler, ein6-1, een-1 and ein6-1 een-1 seedlings. Occupancy of H3K27me3, H2A.Z, H3K4me3 and H2Aub was determined with ChIP-seq, mRNA expression was measured with RNA-seq and levels of methylated cytosines (CG in yellow, CHG in blue, CGG in pink) were determined with MethylC-seq. To ensure an accurate comparison of individual chromatin features between genotypes, the tracks were normalized to the respective sequencing depth. Normalization was separately done for each chromatin feature. Biological replicate 1 of the H3K27me3 and H2A.Z ChIP-seq datasets is shown. (B) Venn diagram illustrates the overlap between genes that show a significant increase of H3K27me3 (2-fold enrichment over ein6-1), H2AZ (2-fold enrichment over een-1) and H2Aub (2-fold enrichment over Ler) in ein6-1 een-1 mutants and also a significant decrease of H3K4me3 (1.5-fold enrichment in Ler over ein6-1 een-1). In addition, genes that contain differentially methylated regions (DMRs) with ten or more methylated cytosines in at least one genotype were included as well. (C) Graphical illustration of H3K27me3 and EIN6 occupancy at the EIN2 gene determined with ChIP-seq. Sequencing reads were merged between biological replicates for the H3K27me3 ChIP-seq using untreated 3-day-old etiolated Ler (gray) and ein6-1 een-1 (blue) seedlings (two replicates each) and the EIN6 ChIP-seq (red) using Ler 35S:EIN6-FLAG seedlings (three replicates). The occupancy was calculated as the ratio between the respective merged ChIP and the merged Ler IgG control in 100 bp bins from 2.4 kb upstream to 7.7 kb downstream of the transcriptional start site (TSS) of EIN2 and is shown as log2 fold change. Negative values which reflect lower occupancy in the ChIP sample compared to the IgG control sample were set to zero.
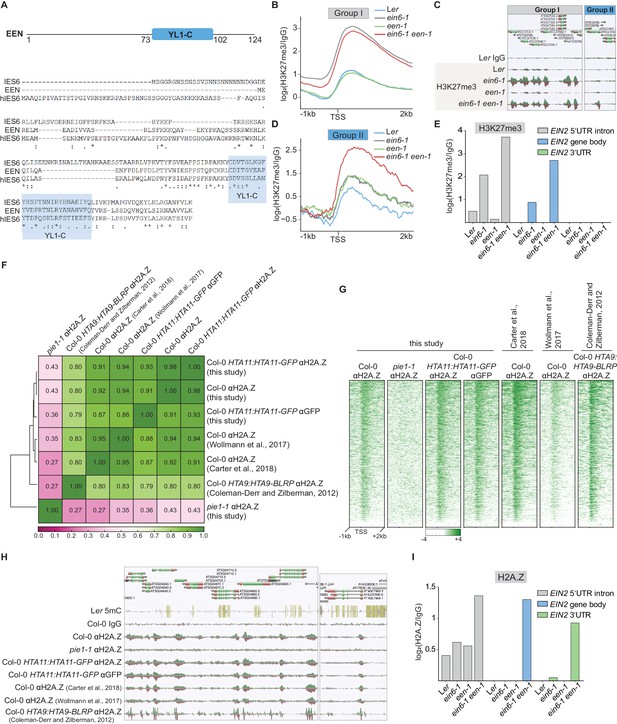
Quantitative description of the chromatin environment at EIN2 in ein6-1 een-1 mutants.
(A) Protein domain structure of EEN as well as a multiple amino acid sequence alignment of EEN, IES6 and human IES6 (hIES6). The YL1-C domain is indicated in blue. (B) Aggregated H3K27me3 profile of 2369 Group I genes shows H3K27me3 occupancy from 1 kb upstream to 2 kb downstream of the TSS in Ler, ein6-1, een-1 and ein6-1 een-1 seedlings. (C) Genome browser screenshot shows differential enrichment of H3K27me3 at an example Group I gene and Group II gene. To ensure an accurate comparison of individual chromatin features between genotypes, the tracks were normalized to the respective sequencing depth. (D) H3K27me3 occupancy of 54 Group II genes in Ler, ein6-1, een-1 and ein6-1 een-1 seedlings is shown as an aggregated H3K27me3 profile from 1 kb upstream to 2 kb downstream of the TSS. The H3K27me3 occupancy was calculated as the ratio between the two respective merged ChIP replicates and the two merged Ler IgG control replicates. (E) Quantification of H3K27me3 levels in the 5’UTR intron, gene body and 3’UTR of the EIN2 gene are shown. The H3K27me3 occupancy in these regions was calculated as the ratio between the respective merged ChIP-seq samples and merged Ler IgG control samples. (F) Spearman’s correlation plot shows correlation of read coverages between the antibody validation H2A.Z datasets from this study (Col-0, pie1-1, Col-0 HTA11:HTA11-GFP (αH2A.Z) and Col-0 HTA11:HTA11-GFP (αGFP)) and three publicly available H2A.Z ChIP-seq datasets (Carter et al., 2018; Wollmann et al., 2017; Coleman-Derr and Zilberman, 2012). Clustering is determined by the degree of correlation. (G) Heatmap shows the H2A.Z occupancy at all Arabidopsis genes in the indicated genotypes. Levels of H2A.Z from 1 kb upstream to 2 kb downstream of the TSS are shown. (H) Genome browser screenshot shows differential enrichment of H2A.Z in Col-0, pie1-1 and Col-0 HTA11:HTA11-GFP seedlings. Moreover, H2A.Z enrichment is also shown for three publicly available H2A.Z ChIP-seq datasets (Carter et al., 2018; Wollmann et al., 2017; Coleman-Derr and Zilberman, 2012). Genetic background and used antibodies are indicated. The Col-0 IgG track serves as a control and the Ler 5mC track indicates methylated cytosines (CG in yellow, CHG in blue, CGG in pink). The shape difference of H2A.Z domains in the Coleman-Derr and Zilberman, 2012 dataset can be explained by the MNase treatment of the chromatin. (I) Levels of H2A.Z in the 5’UTR intron, gene body and 3’UTR of EIN2 in Ler, ein6-1, een-1 and ein6-1 een-1 seedlings are shown. The H2A.Z occupancy in these regions was calculated as the ratio between the respective merged ChIP-seq samples and merged Ler IgG control samples.
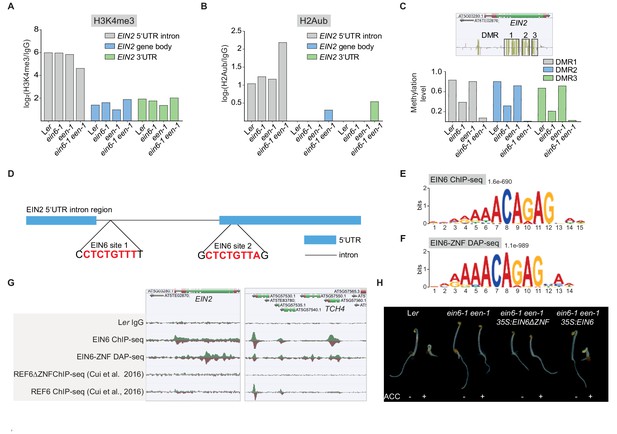
EIN2 displays an aberrant chromatin signature ein6-1 een-1 mutants and is a direct target of EIN6.
(A), (B) Quantification of H3K4me3 (A) and H2Aub (B) levels in the 5’UTR intron, gene body and 3’UTR of EIN2 are shown. The occupancy in these regions was calculated as the ratio between the respective ChIP-seq samples and the Ler IgG control sample. (C) Levels of DNA methylation were detected with MethylC-seq and the three DMRs within the gene body of EIN2 are shown. Exact localization of this three DMR’s (DMR1: chr5, 799482–790672; DMR2: chr5, 791018–791663; DMR3; chr5, 791980–792344) is indicated in a genome browser screenshot showing levels of CG, CHG and CHH at EIN2 in Ler seedlings. (D) Schematic representation indicates the localization of two CTCTGYTY-motifs in the 5’UTR region of EIN2. (E) (F) The CTCTGYTY-motif was discovered as the top-ranked motif in EIN6 ChIP-seq data (E) as well as in EIN6-ZNF DAP-seq data (F) determined by MEME analysis using the 1000 top-ranked peaks that were identified by GEM. (G) Genome browser screenshot visualizes the in vivo binding of full length EIN6 and the in vitro binding of its C-terminal zinc fingers (EIN6-ZNF) to EIN2 and TOUCH4 (TCH4). EIN6-FLAG ChIP-seq and EIN6-ZNF-HALO DAP-seq was employed to monitor in vivo and in vitro binding, respectively. ChIP-seq data from a previous study (Cui et al., 2016) showing, that REF6 but not a truncated REF6 version (REF6ΔZNF) can bind to EIN2 and TCH4, is shown as well. (H) Triple response phenotype of 3-day-old etiolated seedlings of Ler, ein6-1 een-1 and ein6-1 een-1 overexpressing either a wild-type EIN6 version or a truncated version without the zinc fingers (EIN6ΔZNF). Seedlings were grown on control LS media or on LS media supplemented with 10 µM ACC.
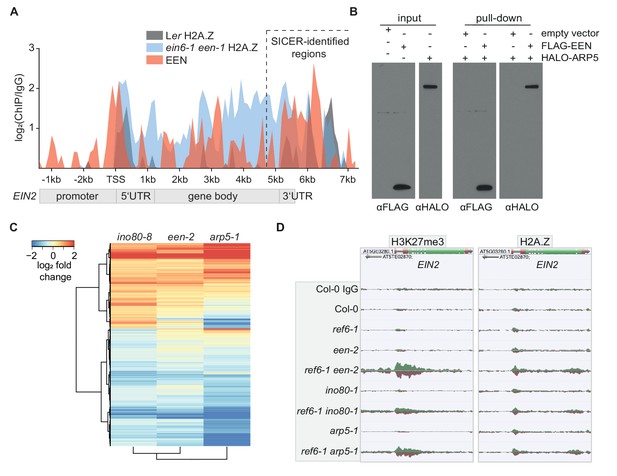
EEN is a subunit of the INO80 chromatin remodeling complex.
(A) Graphical representation of H2A.Z and EEN occupancy at the EIN2 gene determined with ChIP-seq. Sequencing reads of two merged H2A.Z ChIP-seq’s (Ler (gray), ein6-1 een-1 (blue)) and one EEN ChIP-seq (Col-0 35S:EEN-FLAG (red)) using 3-day-old etiolated seedlings were used. The occupancy is displayed as log2 fold change and was calculated as the ratio between the respective ChIP and the respective IgG control in 100 bp bins from 2.4 kb upstream to 7.7 kb downstream of the transcriptional start site (TSS) of EIN2. Occupancies that were lower in the ChIP sample compared to the IgG sample were set to zero. The region with significant EEN enrichment determined with SICER is indicated as well. (B) FLAG-pull-down assays using FLAG-tagged EEN and Halo-tagged ARP5 that were in vitro translated in rabbit reticulocyte extract. Anti-FLAG and anti-Halo antibodies were used to confirm EEN/ARP5 expression, EEN immunoprecipitation and EEN-ARP5 interaction. (C) Heatmap illustrates the log2 fold change in mRNA expression of INO80-dependent genes in untreated 3-day-old etiolated ino80-8, een-2 and arp5-1 seedlings relative to their expression in Col-0 seedlings. INO80-dependent genes were selected as genes that show a significant differential expression in ino80-8 mutant seedlings compared to Col-0 seedlings (787 genes up, 1079 genes down). (D) Genome browser screenshot shows the occupancy of H3K27me3 (right) and H2A.Z (left) at the EIN2 gene in untreated 3-day-old etiolated Col-0, ref6-1, een-2, ref6-1 een-2, ino80-1, ref6-1 ino80-1, arp5-1 and ref6-1 arp5-1 seedlings. Genome-wide occupancy of H3K27me3 and H2A.Z was determined by ChIP-seq. To ensure an accurate comparison of individual chromatin features between genotypes, the tracks were normalized to the respective sequencing depth. Normalization was separately done for each chromatin feature.
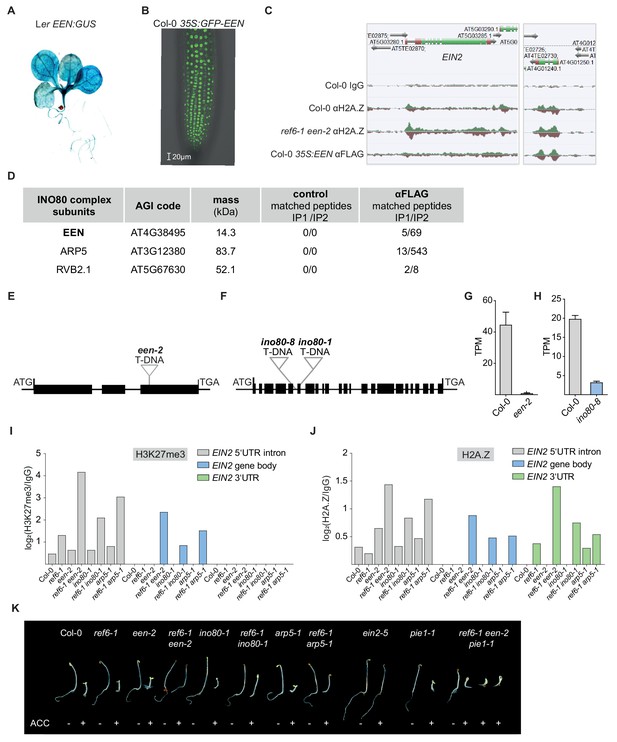
Proteomic and genetic validation of EEN as a subunit of the Arabidopsis INO80 chromatin remodeling complex.
(A) Histochemical localization of GUS expression driven by a 2 kb EEN promoter fragment in 7-day-old Ler EEN:GUS seedlings. (B) GFP fluorescence visualizes subcellular EEN localization in roots of 10-day-old Col-0 35S:GFP-EEN seedlings using confocal laser scanning microscopy. 20 µm scale bar is shown. (C) Genome browser screenshot visualizes the in vivo binding of EEN to the gene body of EIN2, At4g11280 and At4g01250. H2A.Z occupancy in Col-0 and ref6-1 een-2 seedlings at these loci is indicated as well. (D) Mass spectrometry analysis of immuno-purified FLAG-tagged EEN from Col-0 35S:EEN flower tissue revealed ARP5 as an interactor. Results from two independent IPs are shown. Flower tissue of non-transgenic Col-0 was used as a control. Both IPs also detected the helicase RVB2 which is part of the yeast INO80 complex as an interaction partner of EEN. (E), (F) Gene models of EEN and INO80 indicate the localization of the T-DNA insertion in een-2 (E), ino80-1 (F) and ino80-8 (F) mutants. (G), (H) Expression of EEN (G) and INO80 (H) in untreated 3-day-old etiolated Col-0, een-2 and ino80-8 seedlings using RNA-seq is shown. Expression is measured in TPM (Transcripts Per Kilobase Million) and results from two biological replicates are shown (mean ±S.D.). (I) (J) Quantification of H3K27me3 levels (I) and H2A.Z levels (J) in the 5’UTR intron, gene body and 3’UTR of the EIN2 gene in untreated 3-day-old etiolated Col-0, ref6-1, een-2, ref6-1 een-2, ino80-1, ref6-1 ino80-1, arp5-1 and ref6-1 arp5-1 seedlings. Occupancy of H3K27me3 and H2A.Z was determined by ChIP-seq. The H3K27me3 and H2A.Z occupancy in the three regions was calculated as the ratio between the respective ChIP-seq sample and Col-0 IgG control sample. (K) Triple response phenotype of 3-day-old etiolated Col-0, ref6-1, een-2, ref6-1een-2, ino80-1, ref6-1 ino80-1, arp5-1, ref6-1 arp5-1, ein2-5, pie1-1 and ref6-1 een-2 pie1-1 seedlings. Seedlings were grown on control LS media or on LS media supplemented with 10 µM ACC.
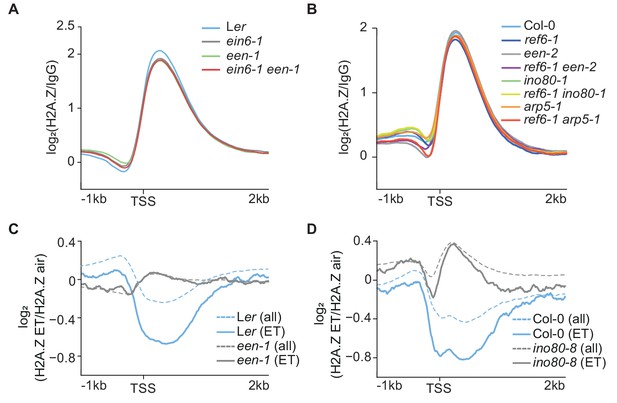
INO80 controls ethylene-induced H2A.Z eviction dynamics.
(A) (B) Aggregated H2A.Z profile of all Arabidopsis genes (TAIR10) in Ler, ein6-1, een-1, ein6-1 een-1 (A) and Col-0, ref6-1, een-2, ref6-1 een-2, ino80-1, ref6-1 ino80-1, arp5-1 and ref6-1 arp5-1 (B) seedlings from 1 kb upstream to 2 kb downstream of the TSS determined by ChIP-seq. (C) (D) ET-induced H2A.Z eviction dynamics are shown as aggregated H2A.Z profiles of 3-day-old etiolated Ler, een-1, Col-0 and ino80-8 seedlings treated for 4 hr with ET gas. Two biological H2A.Z ChIP-seq replicates for Ler and een-1 (C) and one for Col-0 and ino80-8 (D) were analyzed. Profiles visualize the log2 fold change between the respective air and ET-treated H2A.Z ChIP-seq samples from 1 kb upstream to 2 kb downstream of the TSS. 1076 genes that show a robust H2A.Z eviction in response to ET in all replicates (≥1.3 fold SICER comparison, air vs 4 hr ET) are shown as solid lines whereas all genes (TAIR10) are shown as dashed lines.
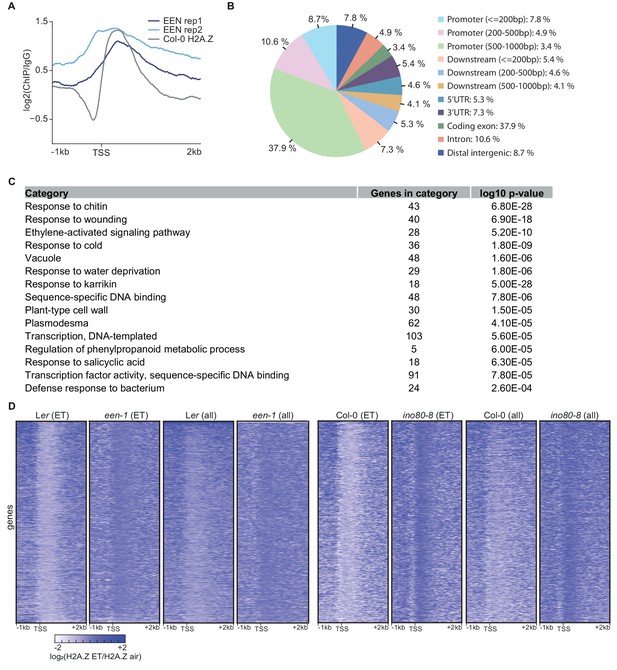
EEN binds to gene bodies of environmentally responsive genes.
(A) Aggregated EEN and H2A.Z profile of the 1000 top-ranked genes that show EEN binding from 1 kb upstream to 2 kb downstream of the TSS. Two independent EEN ChIP-seq datasets (Col-0 35S:EEN) as well as one Col-0 H2A.Z ChIP-seq dataset (Col-0) is shown. (B) Pie chart illustrates the genomic distribution of EEN binding sites. (C) Gene ontology enrichment analysis of the 1000 top-ranked EEN targets using the functional annotation tool DAVID. (D) Heatmaps show ET-induced H2A.Z eviction dynamics in 3-day-old etiolated Ler, een-1, Col-0 and ino80-8 seedlings treated for 4 hr with ET gas. The heatmaps correspond to the aggregate profiles in Figure 4C and D. Heatmaps visualize the log2 fold change between the respective air and ET-treated H2A.Z ChIP-seq samples from 1 kb upstream to 2 kb downstream of the TSS. (ET) indicates that the dynamics of 1076 genes with a robust H2A.Z eviction in response to ET in all replicates (≥1.3 fold SICER comparison, air vs 4 hr ET) is shown whereas (all) indicates that the dynamics of all Arabidopsis genes is shown.
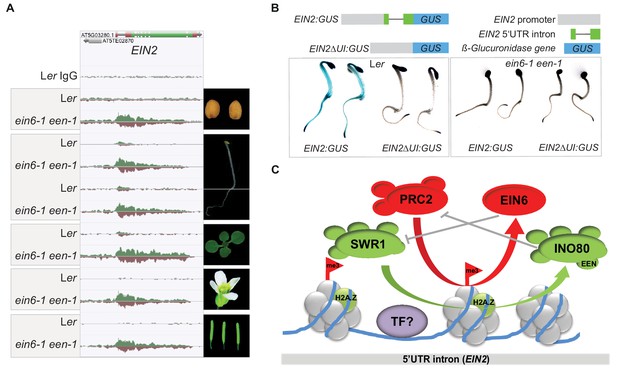
EIN6 (REF6)/INO80-mediated regulation occurs at the 5’UTR intron region of EIN2 during embryogenesis.
(A) Genome browser screenshot visualizes levels of H3K27me3 at the EIN2 gene in the indicated tissues of Ler and ein6-1 een-1 plants (dry seeds, roots and shoots of etiolated seedlings, de-etiolated whole-seedlings, flowers and siliques). H3K27me3 occupancy was identified with ChIP-seq. The tracks were normalized to the respective sequencing depth for each experiment to allow an accurate comparison between Ler and ein6-1 een-1 H3K27me3 profiles. Normalization was separately done for each chromatin feature. (B) Histochemical GUS staining of untreated three-day-old etiolated Ler and ein6-1 een-1 seedlings stably expressing either a EIN2 promoter GUS fusions with (EIN2:GUS) or without the 5’UTR intron (EIN2ΔI:GUS). Schematic illustration of the used EIN2:GUS constructs is shown as well. The gray box indicates the 2 kb promoter region of EIN2, the green boxes represent the 5’UTR and the black line indicates the 5’UTR intron. (C) Hypothetic model of the EIN6 (REF6)/INO80-mediated double safeguard mechanism at the 5’UTR intron of EIN2. Besides the demethylase function role of EIN6 (REF6) in antagonizing PRC2-mediated tri-methylation of H3K27, we speculate that EIN6 (REF6) also antagonizes SWR1-facillitated H2A.Z incorporation at EIN2. This scenario of a dual function holds true for the INO80 complex as well which counteracts SWR1 by removing H2A.Z, but also antagonizes H3K27 trimethylation through an unknown mechanism. The resulting low levels of H3K27me3 and H2A.Z possibly increase the accessibility of the enhancer element for binding of yet unknown transcription factors (TFs). When this EIN6 (REF6)/INO80-mediated regulation is functionally impaired, H3K27me3 and H2A.Z start to accumulate over the entire gene body of EIN2 mutually reinforcing their depositions.
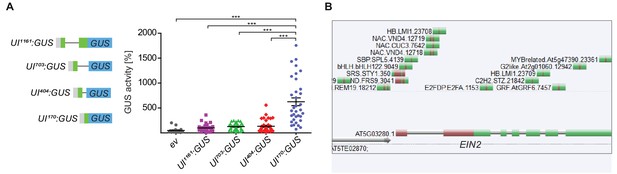
The 5’UTR intron region of EIN2 is crucial for EIN2 expression.
(A) Quantitative GUS assays of Nicotiana benthamiana leaves that transiently express different EIN2 5’UTR intron-driven GUS constructs. Graphic scheme illustrates the full-length and 3’ truncated 5’UTR intron constructs. The gray box indicates truncated promoter region of EIN2, whereas the green boxes and the black line represent the 5’UTR and the intron, respectively. An empty pMDC163 plasmid served as a control (ev). The five constructs were always injected into the same leaf. The average GUS activity of the full length 5’UTR intron construct (UI1161:GUS) was set to 100% in each of the three independent experiments. For UI703:GUS, UI404:GUS and UI170:GUS, the values (±SE) from 34 transformed leaves are shown. Sixteen leaves were used for the empty vector control. Asterisks represent significant differences between UI170:GUS and all other constructs (one-way ANOVA with Bonferroni post test, ***p<0.001). (B) Genome browser screenshot from the Plant Cistrome Database shows TFs that can bind to the 5’UTR region of EIN2 in vitro. Green bars indicate DAP-seq peaks and red bars within them indicate the respective TF-specific binding motif.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (A. thaliana) | ein6-1 een-1 | Roman et al. (1995) | ||
| Strain, strain background (A. thaliana) | ein6-1 | This study | Materials and methods subsection: Plant material and growth conditions | |
| Strain, strain background (A. thaliana) | een-1 | This study | Materials and methods subsection: Plant material and growth conditions | |
| Strain, strain background (A.thaliana) | ein2-45 | Beaudoin et al. (2000) | ||
| Strain, strain background (A. thaliana) | ein2-5 | Alonso et al. (1999) | ||
| Strain, strain background (A. thaliana) | ref6-1 | Noh et al. (2004) | ||
| Strain, strain background (A. thaliana) | een-2 | This study | SALKseq_129237 | Materials and methods subsection: Plant material and growth conditions |
| Strain, strain background (A. thaliana) | arp5-1 | Kandasamy et al. (2009) | ||
| Strain, strain background (A. thaliana) | ino80-1 | Fritsch et al. (2004) | ||
| Strain, strain background (A. thaliana) | ino80-8 | This study | SALKseq_041809 | Materials and methods subsection: Plant material and growth conditions |
| Strain, strain background (A. thaliana) | pie1-1 | Noh and Amasino (2003) | ||
| Strain, strainbackground (A.thaliana) | ref6-1 een-2 | This study | Materials and methods subsection: Plant material and growth conditions | |
| Strain, strain background (A. thaliana) | ref6-1 arp5-1 | This study | Materials and methods subsection: Plant material and growth conditions | |
| Strain, strain background (A. thaliana) | ref6-1 ino80-1 | This study | Materials and methods subsection: Plant material and growth conditions | |
| Strain, strain background (A. thaliana) | pie1-1 ref6-1 een-2 | This study | Materials and methods subsection: Plant material and growth conditions | |
| Strain, strain background (A. thaliana) | Col-0 HTA11:HTA11:GFP | Kumar and Wigge (2010) | ||
| Strain, strain background (A. thaliana) | Ler 35S:EIN6-FLAG | This study | Materials and methods subsection: Vectors construction and plant transformation conditions | |
| Strain, strain background (A. thaliana) | Col-0 35S:FLAG-EEN | This study | Materials and methods subsection: Vectors construction and plant transformation conditions | |
| Strain, strain background (A. thaliana) | Col-0 35S:GFP-EEN | This study | Materials and methods subsection: Vectors construction and plant transformation conditions | |
| Strain, strain background (A. thaliana) | ein6-1 een-1 EIN6:EIN6 | This study | Materials and methods subsection: Vectors construction and plant transformation conditions | |
| Strain, strain background (A. thaliana) | ein6-1 een-1 EEN:EEN | This study | Materials and methods subsection: Vectors construction and plant transformation conditions | |
| Antibody | Rabbit polyclonal anti-histone H3K27me3 | Active Motif | Cat# 39156, RRID:AB_2636821 | 5 µl Materials and methods subsection: ChIP-seq |
| Antibody | Rabbit polyclonal anti - Htz1/Histone H2A.Z | Active Motif | Cat# 39647 RRID:AB_2793289 | 10 µl Materials and methods subsection: ChIP-seq |
| Antibody | Rabbit monoclonal anti trimethyl-Histone H3 (Lys4) | Millipore Sigma | Cat#: 04–745 RRID:AB_1163444 | 4 µl Materials and methods subsection: ChIP-seq |
| Antibody | Rabbit monoclonal anti-ubiquityl-Histone H2A (Lys119) | Cell Signaling Technology | Cat#: 8240 RRID:AB_10891618 | 10 µl Materials and methods subsection: ChIP-seq |
| Antibody | Mouse monoclonal anti-FLAG M2 | Millipore Sigma | Cat#: F1804 RRID:AB_262044 | 5 µl Materials and methods subsection: ChIP-seq |
| Antibody | Mouse monoclonal anti-GFP | Millipore Sigma | Cat#:11814460001 RRID:AB_390913) | 5 µl Materials and methods subsection: ChIP-seq |
| Antibody | ChromPure Mouse IgG, whole molecule | Jackson ImmunoResearch | Cat#:015-000-003 RRID:AB_2337188 | 2 µl Materials and methods subsection: ChIP-seq |
| Antibody | Mouse monoclonal anti-HaloTag | Promega | Cat#: G9211 | 1:1000 Materials and methods subsection: Immunoblot analysis and in vitro pull-down assays |
| Antibody | Rabbit polyclonal anti-Histone H3 | Abcam | Cat#: ab1791 RRID:AB_302613) | 1:1000 Materials and methods subsection: Immunoblot analysis and in vitro pull-down assays |
| Antibody | Rabbit polyclonal anti-EIN3 | Chang et al. (2013) | 1:500 Materials and methods subsection: Immunoblot analysis and in vitro pull-down assays | |
| Antibody | Mouse polyclonal anti-EIN2 | Qiao et al. (2012) | 1:500 Materials and methods subsection: Immunoblot analysis and in vitro pull-down assays | |
| Chemical compound, drug | Protein G Dynabeads | Thermo Fisher Scientific | Cat. #: 10004D | 50 µl Materials and methods subsection: ChIP-seq |
Additional files
-
Supplementary file 1
Differentially regulated genes in Ler, ein6-1, een-1, ein6-1 een-1 and ein2-45 seedlings in response to ET.
Tables displaying differentially regulated ET genes in Ler, ein6-1, een-1, ein6-1 een-1 and ein2-45 seedlings. Differentially expressed genes were discovered with edgeR.
- https://doi.org/10.7554/eLife.47835.013
-
Supplementary file 2
Sequencing samples.
Table displaying all generated sequencing samples.
- https://doi.org/10.7554/eLife.47835.014
-
Supplementary file 3
Group I and Group II genes.
Tables displaying Group I and Group II genes which were identified by SICER through the comparison of ein6-1 vs Ler (Group I) and ein6-1 een-1 vs ein6-1 (Group II).
- https://doi.org/10.7554/eLife.47835.015
-
Supplementary file 4
Differentially enriched epigenome features in ein6-1 een-1 mutants.
Table displaying differentially enriched epigenome features in ein6-1 een-1 mutants. Epigenome feature differences were determined with SICER (H2A.Z, H3K4me3 and H2Aub) and methylpy (5mC).
- https://doi.org/10.7554/eLife.47835.016
-
Supplementary file 5
Binding sites of EIN6, EIN6-ZNF and EEN.
Tables display the binding sites of EIN6, EIN6-ZNF and EEN that were determined with SICER and GEM.
- https://doi.org/10.7554/eLife.47835.017
-
Supplementary file 6
Potential EEN interactors.
Tables display the two IP mass spectrometry replicates using Col-0 35S:EEN-FLAG flowers (MS-IP I and MS IP II). Only potential interactors that show no spectral counts in the IgG sample were included.
- https://doi.org/10.7554/eLife.47835.018
-
Supplementary file 7
Differentially regulated genes in untreated ino80-8, een-2 and arp5-1 seedlings.
Tables displaying differentially regulated genes in ino80-8, een-2 and arp5-1 seedlings. Differentially expressed genes were discovered with edgeR.
- https://doi.org/10.7554/eLife.47835.019
-
Supplementary file 8
Genes with a robust eviction of H2A.Z in response to ET.
Tables displays genes that a robust ET-induced eviction of H2A.Z in Ler and Col-0 (1.3 fold enrichment). Significant differential enrichment was determined with SICER. A list
- https://doi.org/10.7554/eLife.47835.020
-
Supplementary file 9
List of primers.
Table displays all used primer in this study.
- https://doi.org/10.7554/eLife.47835.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47835.022





