Evolutionary emergence of Hairless as a novel component of the Notch signaling pathway
Figures
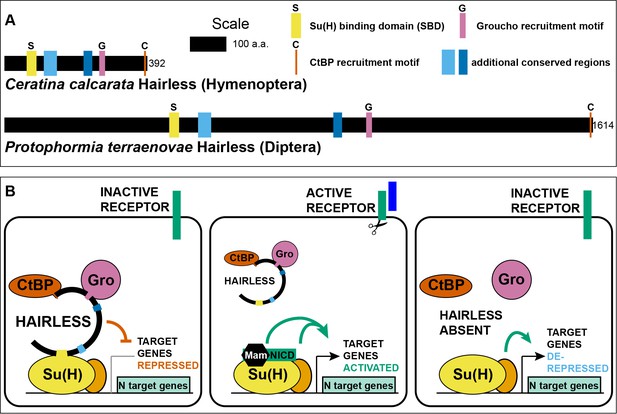
Hairless mediates indirect recruitment of co-repressor proteins to Su(H).
(A) Diagram denoting locations of conserved domains and motifs within Hairless, and illustrating extreme size differences of the protein in different species. Shown is Hairless from the carpenter bee Ceratina calcarata and the blowfly Protophormia terraenovae (Hase et al., 2017), with scale and protein sizes indicated. (B) Summary of Hairless’s known mode of action (Lai, 2002; Maier, 2006) as an adaptor protein that recruits the global co-repressors C-terminal Binding Protein (CtBP) and Groucho (Gro) to Suppressor of Hairless [Su(H)], the transducing transcription factor for the Notch (N) cell-cell signaling pathway; adapted from Figure 6 of Barolo et al. (2002a). In the absence of signaling through the Notch receptor (left), Su(H) acts as a repressor of Notch target genes, despite the presence of transcriptional activator proteins (orange oval). Upon activation of the Notch receptor (middle), Su(H), in a complex with the receptor’s intracellular domain (NICD) and the co-activator Mastermind (Mam), functions to activate transcription of pathway target genes in cooperation with other transcriptional activators. In the absence of Hairless and hence in the absence of Su(H)’s repressive activity (right), the partner transcription factors are often sufficient to activate expression of target genes in a signal-independent manner (Barolo and Posakony, 2002b).
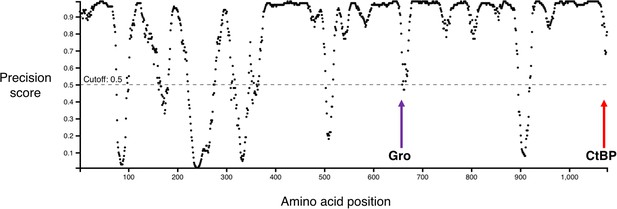
Graph showing predicted disordered regions in Drosophila melanogaster Hairless, generated by DISOPRED3 (Buchan et al., 2013; Jones and Cozzetto, 2015).
Locations of the Gro-binding motif (purple arrow) and CtBP-binding motif (red arrow) are indicated. Note that most of the region between these two motifs is strongly predicted to be disordered.
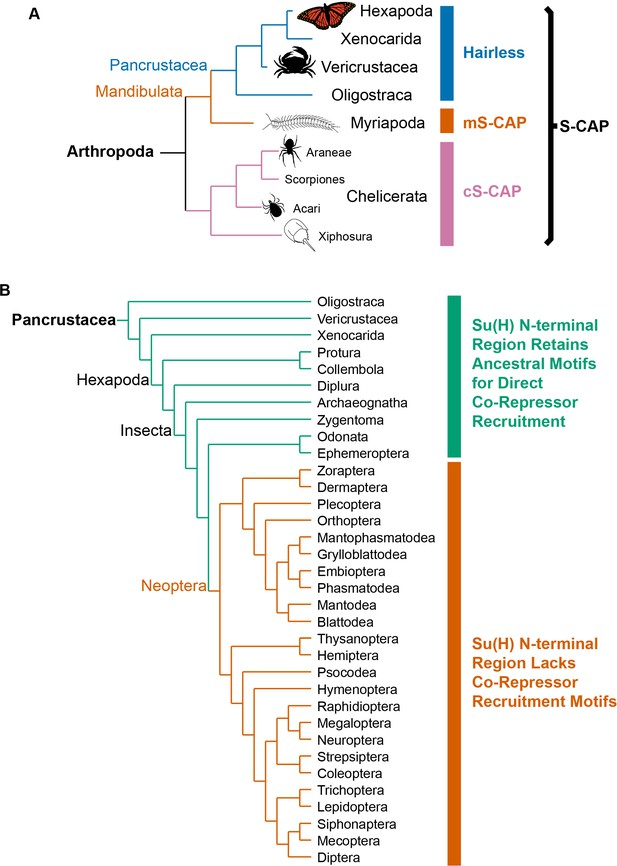
Phylogenetic distribution of Hairless and related S-CAP proteins.
(A) Based on extensive BLAST searches of available genome and transcriptome assemblies, orthologs of the canonical Hairless gene are found only in the Pancrustacea (blue bar), while orthologs of a gene that encodes the related S-CAP protein are found in the Myriapods (mS-CAP, red bar) and Chelicerates (cS-CAP, pink bar). We suggest S-CAP as a suitable umbrella nomenclature for this gene family (black bracket). Tree adapted from Figure 2 of Regier et al. (2010). (B) Consistent with the presence of Hairless as an adaptor protein, Su(H) in most insect orders (the Neoptera clade) has lost the ancestral short linear motifs that mediate direct recruitment of the CtBP and Gro co-repressor proteins (red bar). However, in the Crustacea, Collembola, Diplura, and a subset of Insecta, the ancestral recruitment motifs have been retained in Su(H), despite the presence of Hairless (see Table 1 and Supplementary file 3). Tree adapted from Misof et al. (2014) and Kjer et al. (2016).

Direct binding of co-repressor proteins by Su(H) from the centipede Strigamia maritima.
(A) The PVDLS motif in the N-terminal region of Su(H) from the centipede Strigamia maritima directly binds Drosophila CtBP. A His-tagged 116-aa segment of the Strigamia Su(H) protein, bearing a PVDLS recruitment motif for CtBP, binds strongly to GST-dCtBP (WT, lane 2). Mutation of the motif to alanines (AAAAA) abolishes this interaction (PVDLSmut, lane 1). The results shown in this panel have been replicated in eight additional experiments, utilizing three independent isolations of GST-CtBP protein from bacterial cultures. (B) The conserved GSLTPPDKV motif in the N-terminal region of Strigamia Su(H) directly binds Drosophila Gro. His-tagged E(spl)mβ-HLH protein, which bears a C-terminal WRPW motif that recruits Gro, is used as a binding control. Wild-type (WT) HLHmβ binds GST-Gro (lane 2), while a truncated version of the protein lacking the WRPW motif (lane 1) fails to bind. A synthetic version of HLHmβ in which the WRPW motif has been replaced by the wild-type GSLTPPDKV motif also binds GST-Gro efficiently (lane 4), while a mutant version in which GSLTPPDKV is replaced by alanines (AAAAAAAAA) shows extremely weak binding (lane 3). No binding of any of the His-tagged proteins to GST alone is observed, even with substantially greater amounts of GST compared to GST-Gro. The results shown in this panel have been replicated in seven additional experiments, utilizing five independent isolations of GST-Gro protein from bacterial cultures.
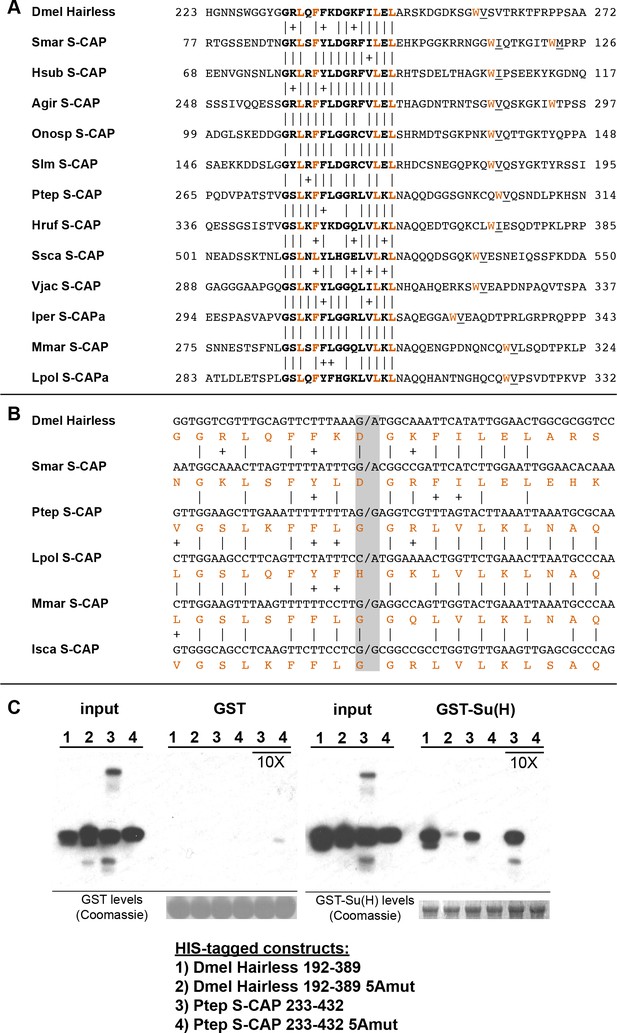
S-CAP proteins in Myriapods and Chelicerates contain a Hairless-like domain that binds Su(H).
(A) Alignment of the Suppressor of Hairless Binding Domain (SBD) in Drosophila melanogaster (Dmel) Hairless with the related motif in the S-CAP proteins from a representative set of Myriapods and Chelicerates. Numbers flanking each sequence segment represent amino acid positions within the protein. The contiguous SBD motif is highlighted in bold. Pairwise amino acid sequence identities within the motifs are indicated by vertical lines; conservative substitutions are indicated by + signs. Amino acids in Hairless that have been shown to make direct contact with Su(H) [including the non-contiguous tryptophan (W) residue] (Yuan et al., 2016) are highlighted in red. Hydrophobic residues nearly always found immediately adjacent to the W are underlined. Species names are as follows: Smar (Strigamia maritima); Hsub (Hydroschendyla submarina) (Fernández et al., 2016); Agir (Anopsobius giribeti) (Fernández et al., 2016); Onosp (Onomeris sp.) (Rodriguez et al., 2018); Slm (Sigmoria latior munda) (Rodriguez et al., 2018); Ptep (Parasteatoda tepidariorum); Hruf (Hypochthonius rufulus) (Bast et al., 2016); Ssca (Sarcoptes scabiei); Vjac (Varroa jacobsoni) (Techer et al., 2019); Iper (Ixodes persulcatus); Mmar (Mesobuthus martensii) (Cao et al., 2013); Lpol (Limulus polyphemus). We note that Maier (2019) has previously described the presence of the SBD-like element in the Strigamia maritima sequence. (B) SBD motifs in both Hairless and S-CAP proteins (red) are encoded in two exons with the same splice junction (indicated by /; see gray highlight). Pairwise amino acid sequence identities within the motifs are indicated by vertical lines; conservative substitutions are indicated by + signs. Species names as in A, except for Isca (Ixodes scapularis). (C) Spider S-CAP protein binds directly to Drosophila Su(H) in vitro. In all panels, lanes 1–4 represent the indicated His-tagged segments of wild-type Drosophila (Dmel) Hairless (lane 1); Dmel Hairless bearing alanine substitutions for each of five SBD residues shown to contact Su(H) (lane 2); wild-type S-CAP from the spider Parasteatoda tepidariorum (Ptep) (lane 3); Ptep S-CAP bearing the same five alanine substitutions (lane 4). Input levels of these His-tagged proteins for each experiment are shown in the respective ‘input’ panels. Remaining two panels show the results of pulldown assays using Sepharose beads bearing only GST (left side) or GST-Su(H) (right side). Left: No binding of the His-tagged proteins to GST alone is observed. Right: Wild-type Dmel Hairless binds efficiently to GST-Su(H) (lane 1); this interaction is severely reduced by the introduction of the five alanine substitutions (lane 2). Wild-type Ptep S-CAP likewise binds to GST-Su(H) (lane 3), while no binding is observed with the alanine-substitution mutant (lane 4); the same result is obtained even when the amount of input Ptep S-CAPs (wild-type and mutant) is increased by a factor of 10 (lanes 3 and 4, 10X). Amounts of GST and GST-Su(H) on the beads are shown in the Coomassie stains below the corresponding pulldown lanes. The results shown in this panel have been replicated in two additional experiments, including one utilizing new isolations of GST-Su(H) and His-tagged proteins.

Sequence characteristics of S-CAP proteins in Myriapods and Chelicerates.
(A) Diagrams of representative examples of Myriapod and Chelicerate S-CAP proteins, denoting locations of SBD motifs and CtBP recruitment motifs. Scale and protein sizes are indicated. (B) Chelicerate, but not Myriapod, S-CAP proteins share N-terminal ELM2 and SANT domains with an MTA zinc-finger protein from the same species. Scale and protein sizes are indicated. (C) Shared ELM2 and SANT domains in Chelicerate MTA and S-CAP proteins are encoded in two exons with the same splice junction (indicated by /; red arrows). Pairwise amino acid sequence identities within the motifs are indicated by vertical lines; conservative substitutions are indicated by + signs. Species names as in Figure 4A.
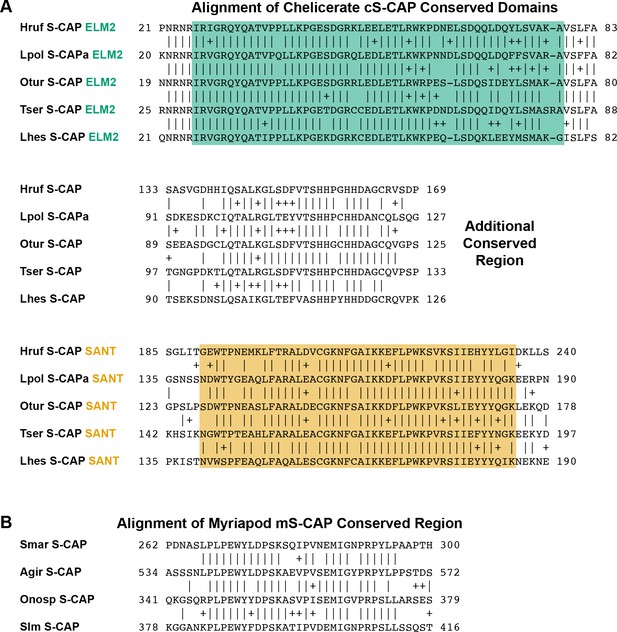
Alignments of sequence regions shared by representative S-CAP proteins from (A) Chelicerates and (B) Myriapods.
Amino acids at the ends of the aligned sequences are numbered. Pairwise amino acid sequence identities are indicated by vertical lines; conservative substitutions are indicated by + signs. ELM2 and SANT domains found in Chelicerate S-CAPs (A) are highlighted in colors corresponding to those in Figure 5B. Species names are as follows: (A) Hruf (Hypochthonius rufulus), Lpol (Limulus polyphemus), Otur (Ornithodoros turicata) (Egekwu et al., 2016), Tser (Tityus serrulatus) (Fuzita et al., 2015), Lhes (Latrodectus hesperus) (Clarke et al., 2015); (B) Smar (Strigamia maritima), Agir (Anopsobius giribeti), Onosp (Onomeris sp.), Slm (Sigmoria latior munda).
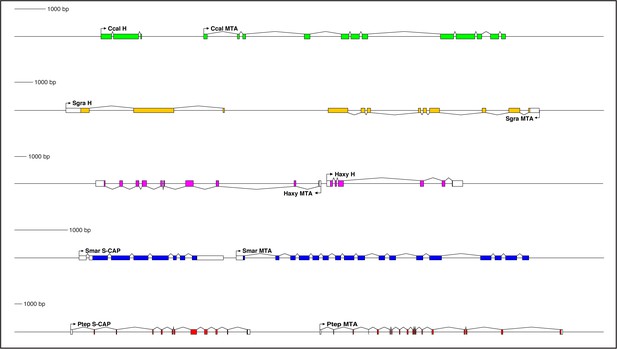
Genes encoding both Hairless and S-CAP proteins are frequently located immediately adjacent to an MTA gene.
Separate scale for each diagram is shown at the left. Three examples are shown for Hairless: the carpenter bee Ceratina calcarata (Ccal), the wheat aphid Schizaphis graminum (Sgra) (QEWZ01001380.1), and the lady beetle Harmonia axyridis (Haxy). Note that microsynteny is often preserved even when gene locations and relative orientations are changed. One example each is shown for S-CAP in Myriapods [the centipede Strigamia maritima (Smar)] and Chelicerates [the house spider Parasteatoda tepidariorum (Ptep)]. See also Supplementary file 1 and Supplementary file 4.
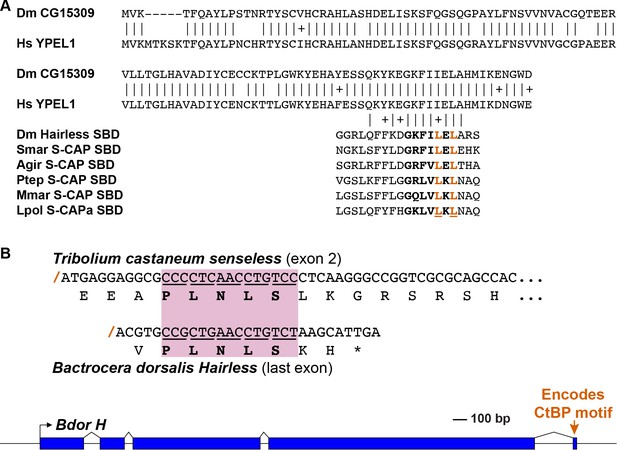
Speculative sources for key elements of the Hairless and S-CAP proteins.
Note that these are intended to be only illustrative examples; other sources are of course possible. (A) A C-terminal segment of the highly conserved Yippee-like protein (Roxström-Lindquist and Faye, 2001; Hosono et al., 2004) is closely related to the C-terminal half of Hairless and S-CAP SBDs. Upper diagram is a sequence alignment of the entire Yippee-like proteins from Drosophila melanogaster (Dm) and Homo sapiens (Hs). Aligned below are contiguous SBD motifs from Drosophila Hairless and five Myriapod and Chelicerate S-CAPs; their C-terminal halves are shown in bold. Two leucine (L) residues shown to make direct contact with Su(H) (Yuan et al., 2016) are highlighted in red. Amino acid sequence identities are indicated by vertical lines; conservative substitutions are indicated by + signs. Other species names as in Figure 4A. (B) As shown in the gene diagram at the bottom, the CtBP recruitment motif in Hairless is encoded by a very small exon located at the extreme 3’ end of the gene [example is from the Oriental fruit fly Bactrocera dorsalis (Bdor; JFBF01000273.1); scale indicated]. A pre-existing gene encoding a protein that utilizes the same PLNLS recruitment motif is a possible source of this exon. Example shown is a portion of the senseless gene from the red flour beetle Tribolium castaneum (Tribolium Genome Sequencing Consortium, 2008). Senseless directly recruits the CtBP co-repressor via the PLNLS motif (Miller et al., 2014). This portion of the protein is encoded in exon 2; splice junction is indicated by a red /. Aligned beneath it is the last exon of the Bdor Hairless gene, illustrating its splice junction in the same frame as senseless exon 2.
Tables
Co-repressor recruitment motifs in protostome Su(H) proteins.
https://doi.org/10.7554/eLife.48115.005| Species | CtBP motif | Gro motif | Source |
|---|---|---|---|
| Ecdyonurus insignis | YPDNHPVDLSSPRPH | APMIPGSLTPPDKMNGEHPHHG | GCCL01029953.1 (Simon et al., 2018) |
| Calopteryx splendens | YTDNHPVDLSSPRPP | HHMIPGSLTPPDKMNGEHPAMH | LYUA01002621.1 (Ioannidis et al., 2017) |
| Atelura formicaria | YPDNHPVDLSSPRPQ | PHMIPGSLTPPDKMNGEHPHHS | GAYJ02050375.1 (Misof et al., 2014) |
| Machilis hrabei | YPDNHPVDLSSPRPH | PHMLPGSLTPPDKMNGEHPHHG | Scaffold 1 (i5K Consortium, 2013) |
| Catajapyx aquilonaris | STANNPVDLSSPRGS | APMIPGSLTPPDKVNGEHHSHH | JYFJ02000853.1 (i5K Consortium, 2013) |
| Holacanthella duospinosa | VPNSNPVDLSNPSPS | SNFVPGSLSPPERMNGNDPSLL | NIPM01000059.1 (Wu et al., 2017) |
| Pollicipes pollicipes | YPDNHPVDLSSPRPE | GPLIAGSLTPPDKLGAELGLHA | GGJN01104381.1 (unpublished) |
| Hyalella azteca | SLGHRPVDLSQAPSP | AAMLAGSLTPPDKLNSDPQQQQ | NW_017238139.1 (i5K Consortium, 2013) |
| Eurytemora affinis | SETSAPVDLSAPRPN | YGMLPGSLTPPDKLNGDHCSPG | NW_019396104.1 (i5K Consortium, 2013) |
| Triops cancriformis | HPEARPVDLSSSRLL | YHSSSLTLTPPDKVNVDGSNSQ | BAYF01001879.1 (Ikeda et al., 2015) |
| Argulus siamensis | YPENNPVDLSNSRTG | SPMIPGSLTPPDKMNGEHHPGH | JW959185.1 (Sahoo et al., 2013) |
| Strigamia maritima | FADNHPVDLSNSHRG | SHMIAGSLTPPDKVNGEHGHQL | JH430541.1 (Chipman et al., 2014) |
| Sigmoria latior munda | TNENHPVDLSSSHRS | SHMIPGSLTPPDKGNAEHSHSH | (Rodriguez et al., 2018) |
| Metaseiulus occidentalis | GADRKPLDMSAAHRS | NW_003805473.1 (Hoy et al., 2016) | |
| Ixodes scapularis | QAAGAPVDMSSHPAR | NW_002722632.1 (Gulia-Nuss et al., 2016) | |
| Parasteatoda tepidariorum 1 | VIDSHPVDLSSPKPS | NW_018383625.1 (Schwager et al., 2017) | |
| Parasteatoda tepidariorum 2 | RYEGRPVDLSSPRPN | NW_018370942.1 (Schwager et al., 2017) | |
| Limulus polyphemus 1 | PYDGHPVDLSNQRPD | NW_013671976.1 (Battelle et al., 2016) | |
| Limulus polyphemus 2 | TYESHPVDLSNQRPD | NW_013676581.1 (Battelle et al., 2016) | |
| Centruroides sculpturatus | GYESSPVDLSSHRSV | MQLISGSMTSHDKVNGDQHSLG | NW_019384406.1 (Schwager et al., 2017) |
| Euperipatoides kanangrensis | NSYDNPVDLSSHRSS | QQILPGSLGPSDKVNGDLVSLA | LN881712.1 (unpublished) |
| Naineris dendritica | DPNGHPVDLSHSRHI | PHMIHGSLTPPDRVNGEPGSGL | (Andrade et al., 2015) |
| Platynereis dumerilii | MASENPVDLSSRHVG | GNHFPGTLTPPDKLNGDHNAHH | KP293861.1 (Gazave et al., 2017) |
| Nephasoma pellucidum | AGYETPVDLSSPRPC | SHLIPGSLTPPDKINGEGITTS | (Lemer et al., 2015) |
| Owenia sp. | QPYENPVDLSRRHIK | AHLIPGSLTPPDKINGDMVTMA | (Andrade et al., 2015) |
| Octopus bimaculoides | NGFDNPMDLSNGKVV | HLMPAGSLTPPDKISGDSISMA | NW_014678436.1 (Albertin et al., 2015) |
| Crassostrea gigas | GGYENPMDLSSNKPG | SHIVAGSLTPPEKINGDPGAMA | NW_011936122.1 (Zhang et al., 2012) |
| Lottia gigantea | AGVENPVDLSNGRIS | SHLFTGSLTPPEKPNGDLVPMS | NW_008708401.1 (Simakov et al., 2013) |
| Notospermus geniculatus | VQYDNPIDLSNRLEG | NHMIPGSLTPPDKVNGDMVPLP | GFRY01035878.1 (Luo et al., 2018) |
| Malacobdella grossa | LHYDNPLDLTNRLDE | GSGIAGSMTPPDGGKGNDLDLQ | (Whelan et al., 2014) |
| Lingula anatina | GGYENPMDLSRRTEM | AHMIPGNLTPPDKVNGEMVPMA | GDJY01029776.1 (Luo et al., 2015) |
| Phoronis australis | QHDNRPMDLSSRGQH | SHLIAGSLTPPDKVNGDVVSMA | GFSC01078935.1 (Luo et al., 2018) |
| Procotyla fluviatilis | ETLFEPLDLRSPIGV | GAKZ01044347.1 (unpublished) | |
| Brachionus koreanus | AKDETPIDLSSKKSK | GBXV02009219.1 (Lee et al., 2015) | |
| Xenoturbella bocki | KRYSAPLNLTVHDKC | DVRVLGRLTPPDKQHVNNDVGA | (Brauchle et al., 2018) |
-
Shown are alignments of short linear amino acid motifs (bold) in the N-terminal region of Su(H) proteins that mediate direct recruitment of the co-repressors CtBP and Gro. Column at right shows the source of the corresponding sequence data, with accession numbers and publication citations indicated.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-HIS G antibody, mouse monoclonal | Invitrogen | CAT#46–1008 (now ThermoFisher CAT#R940-25), RRID:AB_2556557 | 1:5000 dilution |
| Antibody | GOAT anti-mouse HRP, polyclonal | Jackson Immuno-research | CAT#115-035-003, RRID:AB_10015289 | 1:10000 dilution |
| Recombinant DNA reagent | GST-dCtBP pGEX-5X-3 clone | Nibu et al., 1998 | Construct encoding GST-tagged Drosophila CtBP for expression in E. coli | |
| Recombinant DNA reagent | GST-Gro pGEX-KG clone | This paper | Construct encoding GST-tagged Drosophila Groucho for expression in E. coli | |
| Recombinant DNA reagent | GST-Su(H) pGEX-KG clone | Bailey and Posakony, 1995 | Construct encoding GST-tagged Drosophila Su(H) for expression in E. coli | |
| Recombinant DNA reagent | HIS-H192-389 WT pRSET-C clone | This paper | HIS-tagged expression construct encoding amino acids 192–389 of Drosophila Hairless, synthesized by GeneWiz, Inc, and codon-optimized for expression in E. coli | |
| Recombinant DNA reagent | HIS-H192-389 5AMUT pRSET-C clone | This paper | HIS-tagged expression construct encoding amino acids 192–389 of Drosophila Hairless with five alanine substitutions, synthesized by GeneWiz, Inc, and codon-optimized for expression in E. coli | |
| Recombinant DNA reagent | HIS-HLHmBetaSmar WT pRSET-C clone | This paper | HIS-tagged expression construct encoding Drosophila HLHmBeta with the last four amino acids (WRPW) replaced with the sequence GSLTPPDKV | |
| Recombinant DNA reagent | HIS-HLHmBetaSmar MUT pRSET-C clone | This paper | HIS-tagged expression construct encoding Drosophila HLHmBeta with the last four amino acids (WRPW) replaced with the sequence AAAAAAAAA | |
| Recombinant DNA reagent | HIS-HLHmBetaWT pRSET-C clone | This paper | HIS-tagged expression construct encoding full-length Drosophila HLHmBeta | |
| Recombinant DNA reagent | HIS-HLHmBetatrunc pRSET-C clone | This paper | HIS-tagged expression construct encoding Drosophila HLHmBeta with the last four amino acids (WRPW) deleted | |
| Recombinant DNA reagent | HIS-PtepSCAP233-432 WT pRSET-C clone | This paper | HIS-tagged expression construct encoding amino acids 233–432 of Parasteatoda tepidariorum S-CAP, synthesized by GeneWiz, Inc, and codon-optimized for expression in E. coli | |
| Recombinant DNA reagent | HIS-PtepSCAP233-432 5AMUT pRSET-C clone | This paper | HIS-tagged expression construct encoding amino acids 233–432 of Parasteatoda tepidariorum S-CAP with five alanine substitutions, synthesized by GeneWiz, Inc, and codon-optimized for expression in E. coli | |
| Recombinant DNA reagent | HIS-SmarSu(H)ex2-3 WT pRSET-C clone | This paper | HIS-tagged expression construct containing exons 2–3 of Strigamia maritima Su(H), synthesized by GeneWiz, Inc, and codon-optimized for expression in E. coli | |
| Recombinant DNA reagent | HIS-SmarSu(H)ex2-3 mut pRSET-C clone | This paper | HIS-tagged expression construct containing exons 2–3 of Strigamia maritima Su(H) with a PVDLS > AAAAA coding mutation, synthesized by GeneWiz, Inc, and codon-optimized for expression in E. coli | |
| Recombinant DNA reagent | pGEX-5X-3 | Sigma (formerly Amersham; discontinued) | CAT#28-9545-53 | |
| Recombinant DNA reagent | pRSET-C | Invitrogen | CAT#V35120 | |
| Commercial assay or kit | Chem Illumination Reagents | Pierce ECL Western Blotting Substrate | CAT#32209 | |
| Resource, sequence database | NCBI | NCBI | RRID:SCR_006472 | |
| Software, algorithm | NCBI BLAST | NCBI | RRID:SCR_004870 | |
| Software, algorithm | GenePalette | Smith et al., 2017;Rebeiz and Posakony, 2004;http://www.genepalette.org | ||
| Software, algorithm | DNA Strider | Marck, 1988; Douglas, 1995 | ||
| Software, algorithm | BlastStation-Local64 | TM Software, Inc |
Additional files
-
Supplementary file 1
Representative catalog of Hairless proteins in the Pancrustacea, selected from a curated collection of approximately 400 full-length sequences.
Annotated sequences of the entries in this list are provided in Supplementary file 2. In the ‘MTA microsynteny?’ column, + and – in parentheses denote the relative orientation of the Hairless and MTA genes in the genome. References not in the main reference list are provided in Supplementary file 6.
- https://doi.org/10.7554/eLife.48115.012
-
Supplementary file 2
Annotated FASTA file of sequences of the Hairless proteins included in Supplementary file 1.
Characteristic conserved domains and motifs are colored as in Figure 1A.
- https://doi.org/10.7554/eLife.48115.013
-
Supplementary file 3
Annotated FASTA file of sequences of the Su(H) proteins included in Table 1; species for which a full-length Su(H) sequence is not available are omitted here.
Colors denote conserved sequence features. Motifs for direct recruitment of the CtBP and Gro co-repressor proteins (aligned in Table 1) are shown in red and green, respectively. Large region highlighted in orange is the highly conserved body of Su(H), extending from ‘LTREAM’ to ‘YTPEP’.
- https://doi.org/10.7554/eLife.48115.014
-
Supplementary file 4
Representative catalog of S-CAP proteins in the Myriapoda and Chelicerata, selected from a curated collection of approximately 50 full-length sequences.
Annotated sequences of the entries in this list are provided in Supplementary file 5. In the ‘MTA microsynteny?’ column, + and – in parentheses denote the relative orientation of the S-CAP and MTA genes in the genome. The ‘H SBD splice?’ column indicates whether the Su(H)-binding domain (SBD) in the listed protein is encoded by two exons with the same splice junction as in Hairless. If not, the alternative exon structure is indicated. References not in the main reference list are provided in Supplementary file 6.
- https://doi.org/10.7554/eLife.48115.015
-
Supplementary file 5
Annotated FASTA file of sequences of the S-CAP proteins included in Supplementary file 4.
Characteristic domains and motifs are colored as follows: Su(H)-binding domain (SBD), orange; CtBP recruitment motifs, red.
- https://doi.org/10.7554/eLife.48115.016
-
Supplementary file 6
Sequence data references not cited in the main paper text.
- https://doi.org/10.7554/eLife.48115.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48115.018





