Human perivascular stem cell-derived extracellular vesicles mediate bone repair
Figures
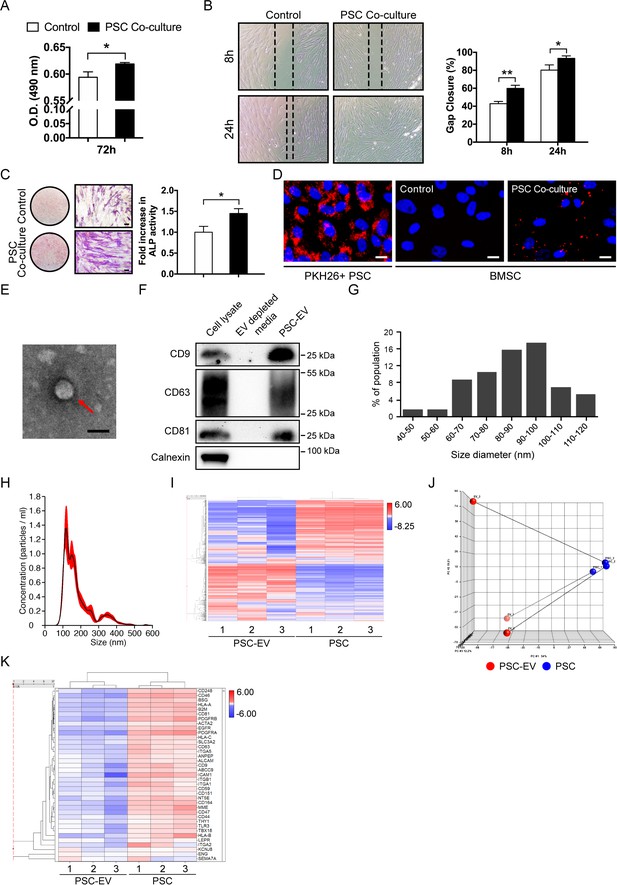
Perivascular stem cells (PSCs) promote BMSC proliferation, migration, and osteogenesis with extracellular vesicle (EV) elaboration in non-contact co-culture.
Human adipose tissue-derived perivascular stem cells (PSCs) were placed in transwell inserts and the effects on human BMSCs were assessed. (A) MTS assay among BMSCs after 72 hr with or without PSCs co-culture. (B) Migration assay among BMSCs after 8 and 24 hr with or without PSCs co-culture. Representative 100x images above, with percentage gap closure below. (C) Alkaline phosphatase staining (left, whole well images and a high magnification view) and quantification (right) among BMSCs after 72 hr with or without PSCs co-culture. (D) BMSCs were cultured with or without PKH26 (red)-labeled PSCs in co-culture. Images (200x) after 48 hr, with DAPI nuclear counterstain (blue). (E) Representative transmission electron microscopy image of PSC-derived extracellular vesicles (PSC-EVs). (F) Western blot for tetraspanin expression (CD9, CD63, and CD81) among PSC cell lysate, EV-depleted supernatant, and purified PSC-EVs. In comparison, the endoplasmic reticulum-associated protein calnexin was also assayed. (G,H) Size distribution of PSC-EVs based on (G) electron microscopy image analysis and (H) Nanoparticle tracking analysis (NanoSight). (I–K) Total RNA sequencing comparison of PSC-EVs to their parent PSCs. (I) Unsupervised hierarchical clustering among PSC-EVs and PSCs. (J) Principal component analysis among PSC-EVs and PSCs. Lines match PSC-EVs to their respective parent cells. (K) Heat map demonstrating mRNA expression levels of putative perivascular markers among PSC-EVs and PSCs. PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem/stromal cell. Data shown as mean ± SD, and represent triplicate experimental replicates in biological duplicate. White scale bar: 20 μm. Black scale bar: 100 nm. *p<0.05; **p<0.01.
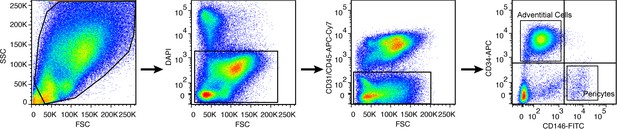
Human PSC derivation by fluorescence activated cell sorting.
From left to right: Perivascular stem cells are derived by size distribution, followed by exclusion of DAPI+ cells, followed by exclusion of CD31 or CD45 expressing cells. Finally, microvascular pericytes (CD146+CD34-) and adventicytes (CD146-CD34+) cells are isolated and combined to constitute a bipartite perivascular stem cell population.
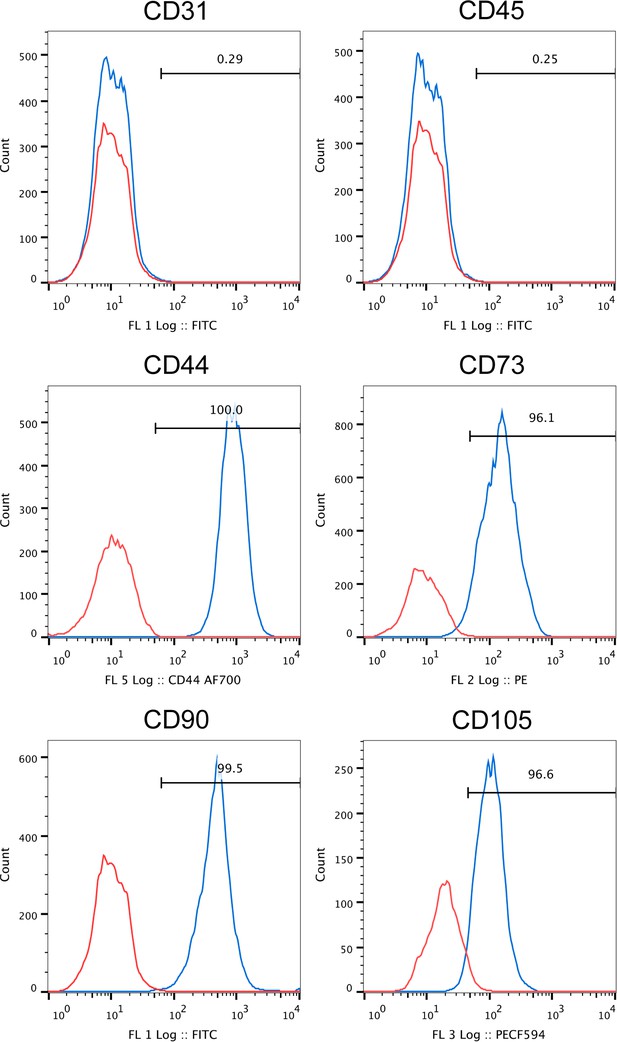
Flow cytometry analysis of FACS derived human PSC.
Human PSC, as defined by FACS purification from human lipoaspirate, were examined at passage eight by flow cytometry. Expression is shown in blue in relation to unstained control in red. No expression of CD31+ endothelial or CD45+ inflammatory cells among FACS purified human PSC. Near universal expression of canonical ‘MSC’ markers among FACS purified human PSC, including CD44, CD73, CD90, and CD105 (frequency: 96.1–100%).
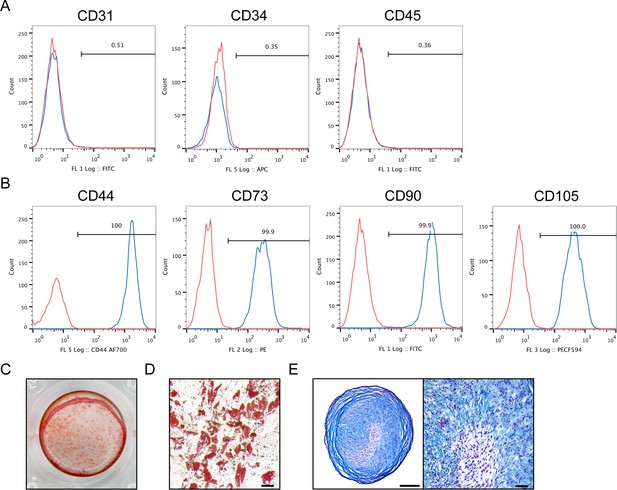
Flow cytometry analysis and multilineage differentiation potential of human culture-derived BMSC.
Human BMSC, as defined by culture-adherence to standard culture ware, were examined at passage two by (A,B) flow cytometry and (C–E) in vitro multilineage differentiation potential. (A,B) By flow cytometry, expression is shown in blue in relation to unstained control in red. (A) From left to right: No expression of CD31+/CD34+ endothelial or CD45+ inflammatory cells among culture expanded human BMSC. (B) Near universal expression of canonical ‘MSC’ markers among culture expanded human BMSC, including CD44, CD73, CD90, and CD105 (frequency: 99.7–100%). (C–E) Multilineage differentiation potential of human BMSC, including (C) osteogenic differentiation as assessed by Alizarin red S staining at d7 of differentiation, (D) adipogenic differentiation as assessed by Oil red O staining at d14 of differentiation (Scale bar: 50 μm), and (E) chondrogenic differentiation as assessed by Alcian blue staining of micromass sections as d21 of differentiation (Left scale bar: 250 μm; right scale bar: 50 μm).
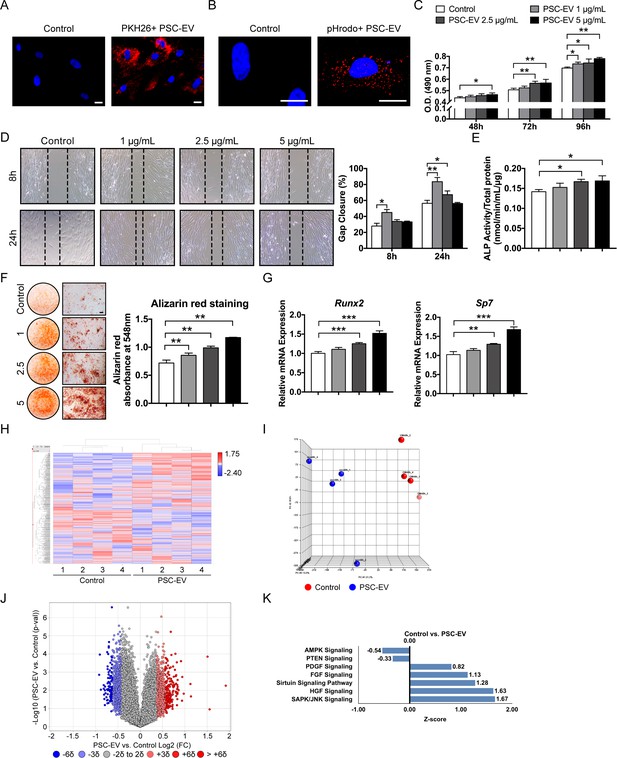
PSC-EVs promote BMSC proliferation, migration, and mineralization.
(A) Appearance of BMSCs treated with or without PKH26 (red)-labeled PSC-EVs. (B) Appearance of BMSCs treated with or without pHrodo (red)-labeled PSC-EVs. Images after 48 hr, with DAPI nuclear counterstain. (C) BMSC proliferation assessed by MTS assay at 48, 72, and 96 hr with or without PSC-EVs (1–5 μg/mL). (D) BMSC migration assessed by scratch wound healing assay at 8 and 24 hr with or without PSC-EVs (1–5 μg/mL). Representative 100x images with percentage gap closure are shown. (E) Alkaline phosphatase (ALP) activity of BMSCs at 3 days in osteogenic medium (OM) treated with or without PSC-EVs (1–5 μg/mL). (F) BMSC osteogenic differentiation assessed by Alizarin Red staining at 7 days in osteogenic medium (OM) with or without PSC-EVs (1–5 μg/mL). Photometric quantification shown right. (G) BMSC osteogenic gene expression by qPCR at 7 days in OM with or without PSC-EVs (1–5 μg/mL), including Runt related transcription factor 2 (Runx2) and Sp7 (Osterix). (H–K) PSC-EVs induced changes in the BMSC transcriptome, as assessed by Clariom D microarray. BMSCs were cultured in the presence or absence of PSC-EVs (2.5 μg/mL) for 48 hr. (H) Unsupervised hierarchical clustering among BMSCs treated with or without PSC-EVs. (I) Principal component analysis among BMSCs treated with or without PSC-EVs. (J) Volcano plot of all transcripts. X-axis represents Log2 fold change for each gene. Y-axis represents -Log10 p value. Red dots indicate >2 SD increase among PSC-EV-treated samples. Blue dots indicate >2 SD decrease among PSC-EV-treated samples. (K) Ingenuity Pathway Analysis (IPA) identified representative pathways that were upregulated (Z-score >0) or downregulated (Z-score <0) among BMSCs treated with PSC-EVs. PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem cell. Data shown as mean ± SD, and represent triplicate experimental replicates. White scale bar: 20 μm. Black scale bar: 100 nm. *p<0.05; **p<0.01; ***p<0.001.
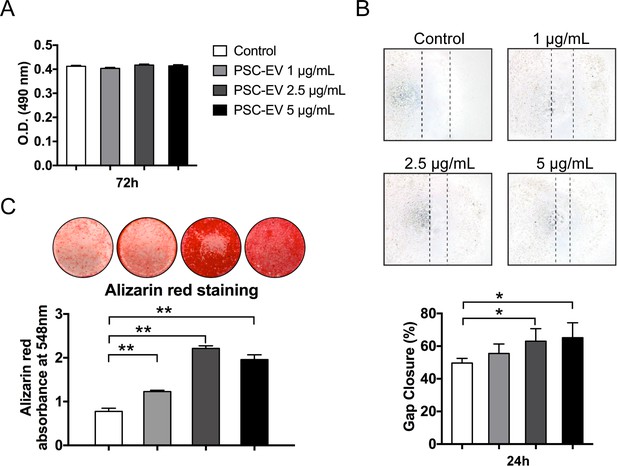
PSC-EV promote ASC migration and mineralization.
In similarity to studies in human BMSC, human unpurified adipose-derived stem stromal cells (ASC) were incubated with PSC-EV at ascending dosages. (A) ASC proliferation assessed by MTS assay at 72 hr with or without PSC-EV (1–5 μg/mL). (B) ASC migration assessed by scratch wound healing assay at 24 hr with or without PSC-EV (1–5 μg/mL). Representative 100x images (above) with percentage gap closure shown (below). (C) ASC osteogenic differentiation assessed by Alizarin Red staining at 7 days in osteogenic medium (OM) with or without PSC-EV (1–5 μg/mL). Photometric quantification shown below. PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle; ASC, adipose-derived stem/stromal cell. Data shown as mean ± SD, and represent triplicate experimental replicates. *p<0.05; **p<0.01.
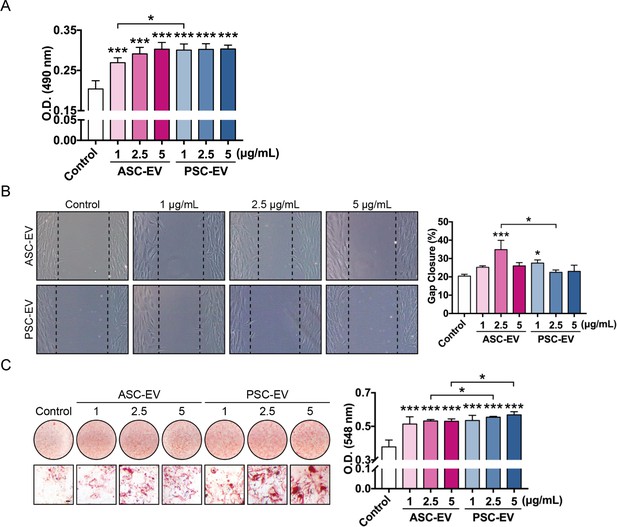
Comparative effects of human ASC-EVs and PSC-EVs on BMSC proliferation, migration, and osteogenesis.
(A) BMSC proliferation assessed by MTS assay at 72 hr with or without ASC-EVs and PSC-EVs (1–5 μg/mL). (B) BMSC migration assessed by scratch wound healing assay at 8 hr with or without ASC-EVs and PSC-EVs (1–5 μg/mL). Representative 100x images with percentage gap closure are shown. (C) BMSC osteogenic differentiation assessed by Alizarin Red staining at 7 days in osteogenic medium (OM) with or without ASC-EVs and PSC-EVs (1–5 μg/mL). Photometric quantification shown right. Scale bar: 50 μm. ASC: adipose-derived stem cell; PSC: perivascular stem cell; ASC-EV: adipose tissue stem/stromal cell-derived extracellular vesicle; PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem cell. Data shown as mean ± SD, and represent triplicate experimental replicates. *p<0.05; **p<0.01; ***p<0.001.
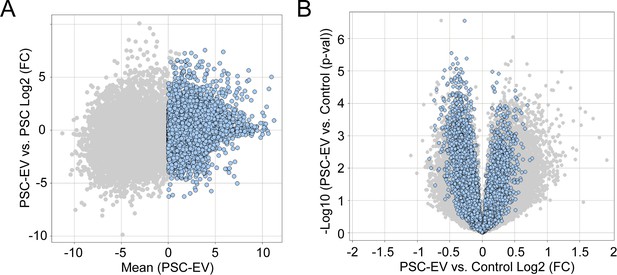
Distribution of PSC-EV associated transcripts within control- or EV-treated BMSC.
(A) Identification of transcripts within PSC-EV. Minus versus average (MvA) plots of all transcripts in RNA sequencing of three PSC-EV samples. Blue dots indicate 7,789 PSC-EV transcripts with a mean FPKM >0. (B) Next, the distribution of these 7,789 PSC-EV transcripts were examined among recipient BMSC 48 hr after treatment with PSC-EV or control. Volcano plot of PSC-EV-treated versus control-treated BMSC. All transcripts are shown in light grey, while the 7,789 PSC-EV transcripts are shown in blue. Of these, approximately half were over-represented in PSC-EV treated cells (47.22%, 3678 transcripts). PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle.
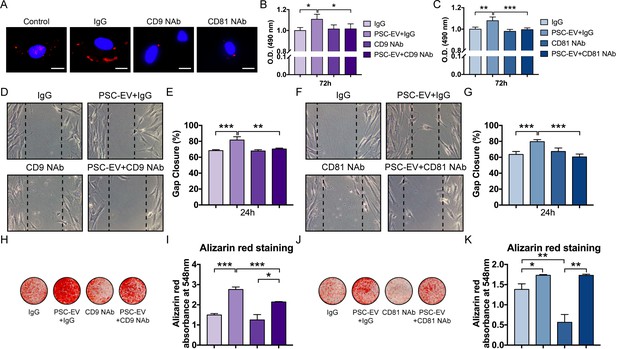
PSC-EVs require tetraspanins for bioactive effects on BMSCs.
(A) Appearance of BMSCs treated with pHrodo (red)-labeled PSC-EVs in the context of incubation with neutralizing antibodies to CD9, CD81, or isotype control (IgG). Images after 48 hr, with DAPI nuclear counterstain. White scale bar: 15 μm. (B,C) BMSC proliferation assessed by MTS assay at 72 hr, with or without anti-CD9 (B) or anti-CD81 (C) neutralizing antibodies. (D,E) BMSC migration assessed by scratch wound healing assay at 8 hr with or without anti-CD9 neutralizing antibodies, shown by microscopic images (D) and quantification (E). (F,G) BMSC migration assessed by scratch wound healing assay at 8 hr with or without anti-CD81 neutralizing antibodies, shown by microscopic images (F) and quantification (G). (H,I) BMSC osteogenic differentiation with or without anti-CD9 neutralizing antibodies, as assessed by Alizarin Red staining (H) and photometric quantification (I) at 7 days in OM. (J,K) BMSC osteogenic differentiation with or without anti-CD81 neutralizing antibodies, as assessed by Alizarin Red staining (J) and photometric quantification (K) at 7 days in OM. PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem cell. Data shown as mean ± SD, and represent triplicate experimental replicates. *p<0.05; **p<0.01; ***p<0.001.
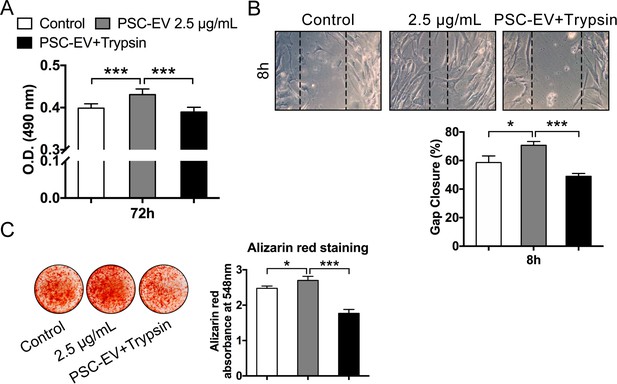
PSC-EVs require surface-associated proteins for bioactive effects on BMSCs.
Trypsinization effect on PSC-EV bioactivity. PSC-EVs were pre-treated with trypsin, followed by re-isolation of EVs and application to BMSCs. (A) BMSC proliferation assessed by MTS assay at 72 hr with PSC-EVs (2.5 μg/mL) with or without trypsinization. (B) BMSC migration assessed by scratch wound healing assay at 8 hr with PSC-EVs (2.5 μg/mL) with or without trypsinization. (C) BMSC osteogenic differentiation assessed by Alizarin Red staining and photometric quantification at 7 days in osteogenic medium (OM) with PSC-EVs (2.5 μg/mL) with or without trypsinization. PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem cell. Data shown as mean ± SD, and represent triplicate experimental replicates. *p<0.05; ***p<0.001.
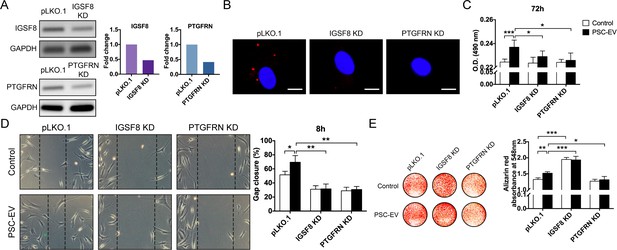
PSC-EV bioactivity requires IGSF8 and PTGFRN expression on recipient BMSCs.
(A) Gene expression of either IGSF8 or PTGFRN as assessed by regular PCR after shRNA mediated knockdown (96 hr shown). (B) Appearance of IGSF8 or PTGFRN shRNA silenced BMSCs treated with pHrodo (red)-labeled PSC-EVs. Images after 48 hr, with DAPI nuclear counterstain. White scale bar: 15 μm. (C) BMSC proliferation assessed by MTS assay at 72 hr with or without IGSF8 or PTGFRN shRNA, with or without PSC-EV treatment (2.5 μg/mL). (D) BMSC migration assessed by scratch wound healing assay at 8 hr with or without IGSF8 or PTGFRN shRNA, with or without PSC-EV treatment (2.5 μg/mL). (E) BMSC osteogenic differentiation assessed by Alizarin Red staining and photometric quantification at 7 days in OM. Data shown as mean ± SD, and represent triplicate experimental replicates. PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem cell. *p<0.05; **p<0.01; ***p<0.001.
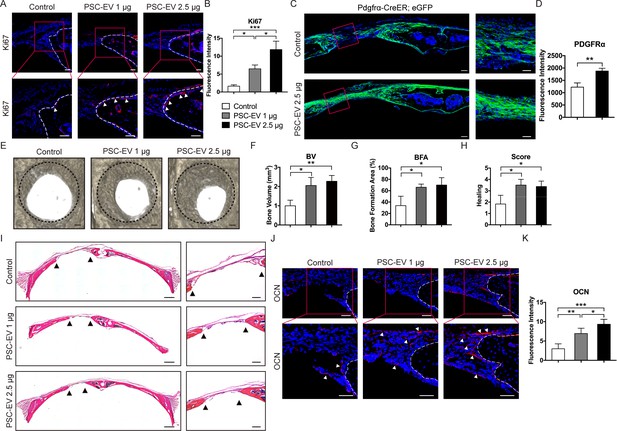
PSC-EVs promote calvarial bone regeneration in vivo.
PSC-EVs (1 or 2.5 μg) were percutaneously delivered twice weekly overlying a circular, full thickness frontal bone defect site (1.8 mm diameter). Analysis was performed at 4 weeks thereafter. See also Figure 5—figure supplement 2 for a schematic representation. (A) Cell proliferation at the bone defect edge (white arrowheads), as assessed by Ki67 immunofluorescent detection. White scale bar: 50 μm. (B) Quantification of bone-lining Ki67 immunoreactivity was shown. (C) Stromal/osteoprogenitor cell migration, as assessed by Pdgfrα-CreER; eGFP cell lineage tracing. Tile scan (left, scale bar: 100 μm) and high-magnification images of the defect site (right, scale bar: 50 μm) demonstrate migration of GFP+ progenitor cells into the defect site. (D) Quantification of eGFP reporter activity in the mid-defect was shown. (E) Bone healing assessed by microcomputed tomography (μCT), shown in a top-down view. Black scale bar: 200 μm. (F–H) μCT analysis including (F) Bone Volume (BV), (G) bone formation area (BFA), and (H) healing score for the extent of bony bridging and bone union. (I) H&E appearance of the defect site. Tile scan (left, scale bar: 500 μm) and high-magnification images of the defect site (right, scale bar: 200 μm), with bone edges indicated by black arrowheads. (J) Osteocalcin (OCN) immunofluorescent detection at the bone defect edge. White scale bar: 50 μm. (K) Quantification of bone-lining OCN immunoreactivity was shown. PSC-EV: perivascular stem cell-derived extracellular vesicle; BMSC, bone marrow mesenchymal stem cell. Data shown as mean ± SD, and represent N = 4 defects per treatment group. *p<0.05; **p<0.01; ***p<0.001.
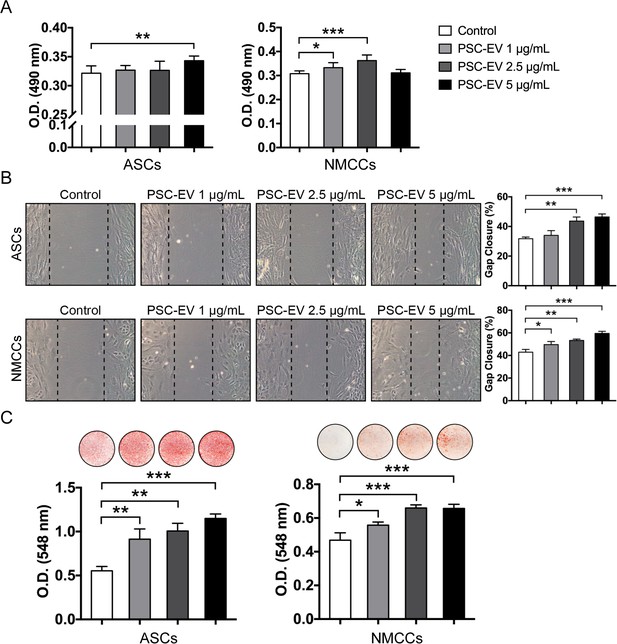
PSC-EVs promote mouse cell proliferation, migration, and mineralization.
Effects on PSC-EV were assayed using either mouse ASCs or neonatal mouse calvarial cells (NMCCs). (A) Mouse ASC and NMCC proliferation assessed by MTS assay at 72 hr with or without PSC-EVs (1–5 μg/mL). (B) Mouse ASC and NMCC migration assessed by scratch wound healing assay at 24 hr with or without PSC-EVs (1–5 μg/mL). Representative 100x images with percentage gap closure are shown. (C) Mouse ASC and NMCC osteogenic differentiation assessed by Alizarin Red staining at 7 days and 12 days, respectively, in osteogenic medium (OM) with or without PSC-EVs (1–5 μg/mL). Photometric quantification shown below. ASC: adipose-derived stem cell; NMCC: Neonatal mouse calvarial cell; PSC: perivascular stem cell; PSC-EV: perivascular stem cell-derived extracellular vesicle. Data shown as mean ± SD, and represent triplicate experimental replicates. *p<0.05; **p<0.01; ***p<0.001.
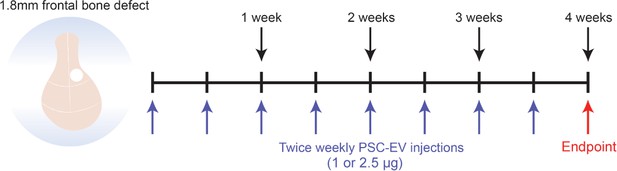
Animal treatment protocol.
Schematic representation of animal treatment protocol for in vivo study. A full-thickness frontal bone defect is created (1.8 mm diameter circular defect in the right frontal bone), followed by percutaneous treatment with PSC-EV (twice weekly for a four week period). The study endpoint was after 4 weeks, at which time postmortem studies were performed.
Additional files
-
Supplementary file 1
Key Resources Table.
- https://doi.org/10.7554/eLife.48191.017
-
Supplementary file 2
Frequency of human PSC in lipoaspirate used.
- https://doi.org/10.7554/eLife.48191.018
-
Supplementary file 3
Yields of PSC-EV.
Yield produced by each PSC-EV isolation are summarized. Protein amounts of harvested PSC-EV were determined by the BCA method. For patient samples 1 and 2, the same cell population was used to isolate EVs at two different passages as indicated.
- https://doi.org/10.7554/eLife.48191.019
-
Supplementary file 4
Highest 100 transcripts in human PSC-EVs.
- https://doi.org/10.7554/eLife.48191.020
-
Supplementary file 5
Relative gene expression within human PSC-EVs or parent PSC among transcription factors enriched in porcine ASC-EVs.
- https://doi.org/10.7554/eLife.48191.021
-
Supplementary file 6
Most upregulated pathways among PSC-EV-treated BMSC by Ingenuity Pathway Analysis.
- https://doi.org/10.7554/eLife.48191.022
-
Supplementary file 7
Most downregulated pathways among PSC-EV-treated BMSC by Ingenuity Pathway Analysis.
- https://doi.org/10.7554/eLife.48191.023
-
Supplementary file 8
CD markers enriched in PSC-EVs.
- https://doi.org/10.7554/eLife.48191.024
-
Supplementary file 9
Animal allocation and treatment groups.
- https://doi.org/10.7554/eLife.48191.025
-
Supplementary file 10
Antibodies used.
- https://doi.org/10.7554/eLife.48191.026
-
Supplementary file 11
Quantitative PCR primers used.
- https://doi.org/10.7554/eLife.48191.027
-
Supplementary file 12
Basic features of human PSCs, ASCs, and BMSCs.
- https://doi.org/10.7554/eLife.48191.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48191.029





