Extensive intraspecies cryptic variation in an ancient embryonic gene regulatory network
Figures
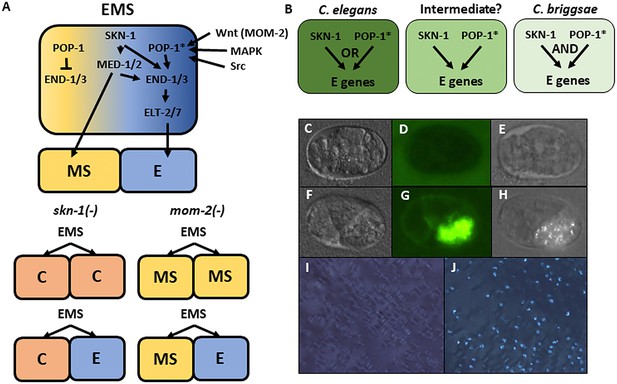
Endoderm regulatory pathway and scoring of gut differentiation.
(A) Under normal conditions, signaling from the posterior P2 cell (Wnt, MAPK and Src) results in asymmetric cortical localization of Wnt signaling pathway components in EMS leading to POP-1 asymmetry in the descendants of EMS, with high levels of nuclear POP-1 in anterior MS and low levels of nuclear POP-1 in the posterior, E, daughter cell. In the anterior MS cell, high nuclear POP-1 represses the END genes, allowing SKN-1 to activate MS fate. In the posterior E cell, which remains in contact with P2, POP-1 is converted to an activator and, along with SKN-1, activates the END genes, resulting in endoderm fate. Loss of skn-1, either by RNAi or in loss-of-function mutants, causes 100% of the embryos to arrest; in 70% of the arrested embryos, EMS gives rise to two C-like cells, while in the remaining 30% only MS is converted to a C fate; the posterior daughter retains its E fate. Loss of mom-2 leads to 100% embryonic arrest with a partially penetrant E→MS cell fate transformation, resulting in two MS-like daughter cells in ~72% of the embryos. (B) Regulatory logic of SKN-1 and POP-1 in E specification in C. elegans, C. briggsae and a hypothetical intermediate state. POP-1* denotes the activated state. (C-H) Gut visualization in embryos affected by skn-1 RNAi. (C-E) arrested embryos without endoderm, (F-H) arrested embryos with endoderm. (C, F) DIC images of arrested embryos ~ 12 hr after egg laying. (D, G) the same embryos expressing the gut-specific elt-2::GFP reporter, and (E,H) birefringent gut granules under polarized light. All embryos showing gut birefringence also show elt-2::GFP expression. (I, J) Fields of arrested skn-1(RNAi) embryos in wild isolate strains JU1491 (I) and JU440 (J), which reflect the extremes in the spectrum of requirement of SKN-1 in gut development at 0.9% and 60%, respectively.
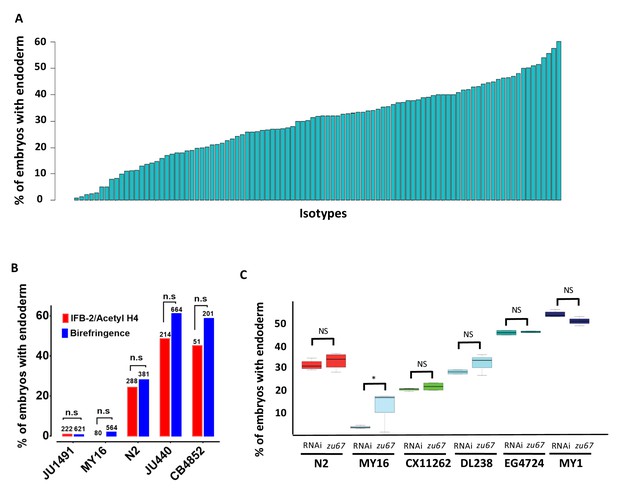
Quantitative effects of loss of skn-1 on endoderm formation.
(A) Spectrum of skn-1(RNAi) effects across the C. elegans isolates. The effects of skn-1(RNAi) are quantified as the average percentage of arrested embryos with endoderm (y-axis). All wild isolates treated with skn-1(RNAi) resulted in 100% embryonic arrest (n > 500 embryos per replicate per isotype and at least two replicates per isotype). (B) Comparison of skn-1(RNAi) phenotype using two different gut markers (birefringent gut granules and MH33 staining of IFB-2) in five different genetic backgrounds. In all cases, no significant statistical difference was found between the two quantitative methods. Fisher’s exact test (NS p-value>0.05). (C) Comparison of skn-1(RNAi) and skn-1(zu67) effects on endoderm development in six different genetic backgrounds. For each color-coded strain, the first value is of the skn-1(RNAi) results (five replicates), while the second is the result for the skn-1(zu67) allele introgression (10 replicates). For all strains (with the exception of MY16), no significant statistical difference was found between the RNAi knockdown and corresponding skn-1(zu67) allele effects on endoderm development. Student t-test (NS p-value>0.05, * p-value<0.05).
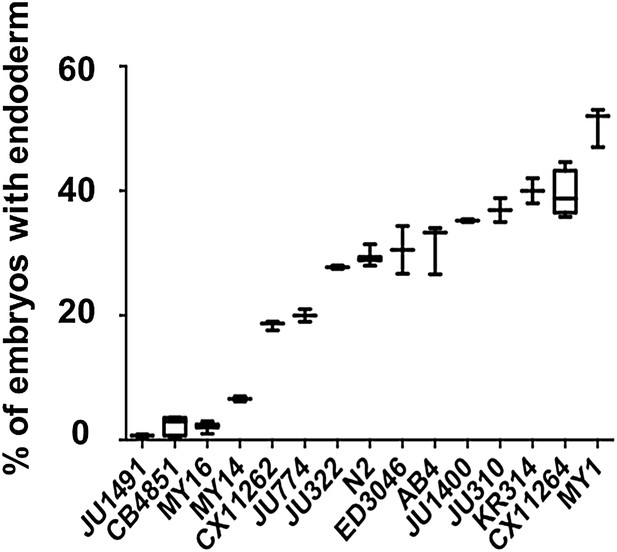
High reproducibility of skn-1(RNAi) phenotypes in various C. elegans isotypes.
elegans isotypes. A minimum of two replicates were obtained, with >500 embryos per replicate (N = 3). Boxplot represents median with range bars showing upper and lower quartiles.
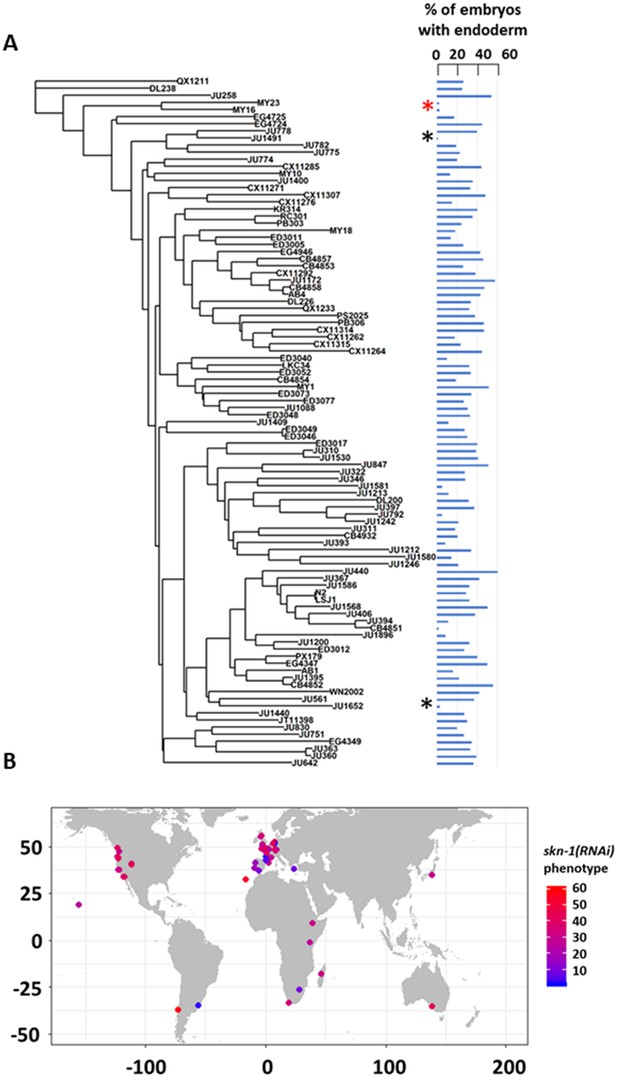
SKN-1 requirement does not correlate with genotypic relatedness or geographical location.
(A) skn-1(RNAi) phenotype of 97 isolates arranged with respect to the neighbor-joining tree constructed using 4,690 SNPs and pseudo-rooted to QX1211. Red asterisk indicates an example of closely related strains (MY23 and MY16) with similar phenotype, while black asterisks indicate example sister strains (JU778 and JU1491; JU561 and JU1652) with distinct phenotype. Phylogenetic relatedness and phenotype are not significantly correlated (Pagel’s λ = 0.42, p-value=0.14). (B) Worldwide distribution of skn-1(RNAi) phenotype across 97 wild isolates. Each circle represents a single isotype.
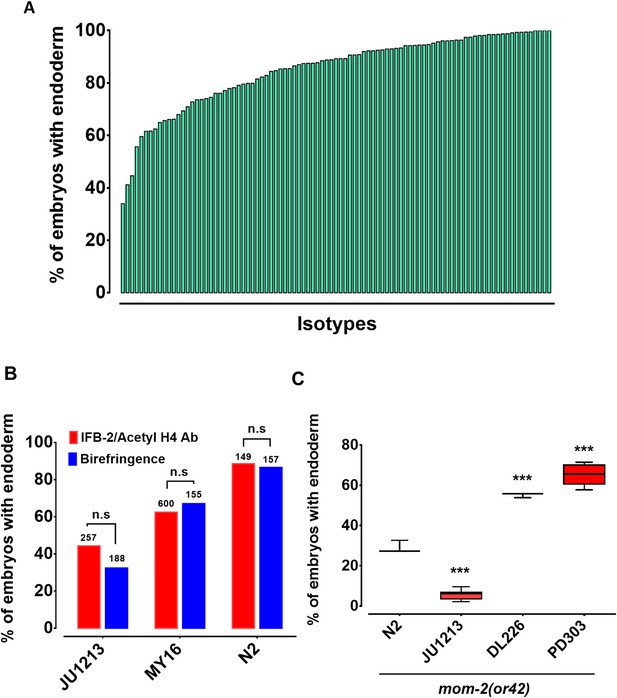
Widespread variation in the mom-2(RNAi) phenotype.
(A) Spectrum of mom-2(RNAi) effects across the C. elegans isolates. The effects of mom-2(RNAi) are quantified as the average percentage of arrested embryos with endoderm (y-axis). Each column represents the mean for each wild isolate (n > 500 embryos were scored for each experiment with at least two replicates per isotype). (B) Comparison of mom-2(RNAi) phenotype using two different gut markers (birefringent gut granules and MH33 immunostaining of IFB-2) in three different genetic backgrounds. In all cases, no significant statistical difference was found between the two quantitative methods. Fisher’s exact test (NS p-value>0.05). (C) Comparison of the effect of mom-2(or42) on endoderm development after introgression into four different genetic backgrounds. At least three independent introgressed lines were studied for each wild isotype. The results were compared to N2; mom-2(or42). Student t-test (*** p-value<0.001).
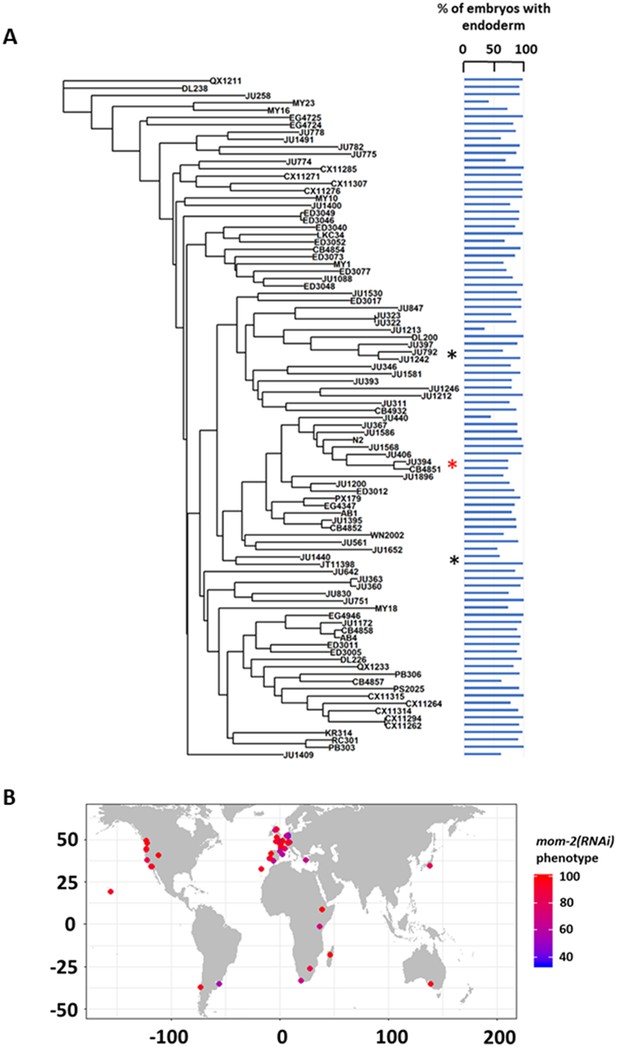
MOM-2 requirement does not correlate with genotypic relatedness or geographical location.
(A) mom-2(RNAi) phenotype of 94 isolates arranged with respect to the neighbor-joining tree constructed using 4,690 SNPs and pseudo-rooted to QX1211. Red asterisk indicates an example of closely related strains (JU394 and CB4851) with similar phenotypes, while black asterisks indicate examples sister strains (JU792 and JU1242; JU1440 and JT11398) with distinct phenotypes (λ = 6.94×10^−05, p-value=1). (B) Worldwide distribution of mom-2(RNAi) phenotype across 94 isolates. Each circle represents a single isolate.
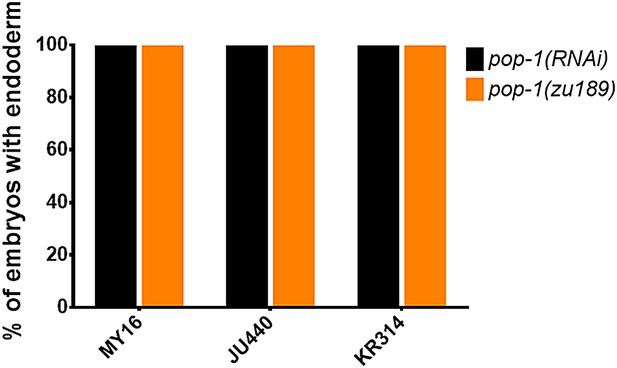
The requirement for POP-1 in endoderm formation does not vary in three introgressed strains.
Strains are shown on the x-axis and fraction of arrested embryos with endoderm are shown on the y-axis. The orange bars represent the results from mutant lines: MY16;pop-1(zu189), JU440;pop-1(zu189), and KR314;pop-1(zu189). Four introgressed lines were created for each new mutant strain. Black bars represent pop-1(RNAi) results on the wild isolates indicated. >200 embryos were scored per experiment.
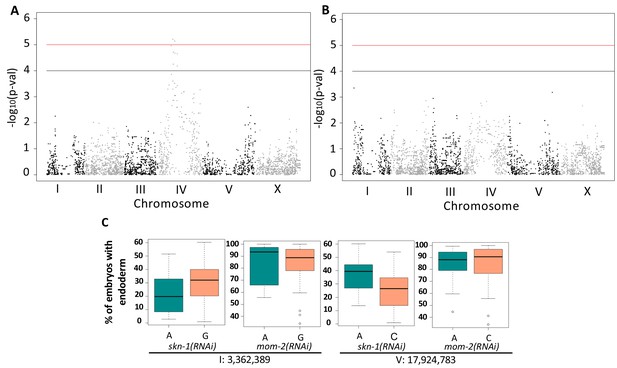
Genome-Wide Association Studies of skn-1(RNAi) and mom-2(RNAi) phenotypes.
(A) Manhattan plot of skn-1(RNAi) GWAS. The red line indicates a genome-wide 1.5% FDR (permutation-based FDR, from 10,000 permutated results). Black line represents 3.0% FDR. The y axis is the –log10 of p-value. (B) Manhattan plot of mom-2 (RNAi) EMMA. The y axis is the –log10 of p-value. Genomic regions are shown on the x-axis. (C) Effect plots of the most strongly-linked SNPs from mom-2(RNAi) GWAS at position 3,362,389 bp on chromosome I and position 17,924,783 bp on chromosome V. Horizontal lines within each box represent the median, and the boxes represent 25th–75th percentile.
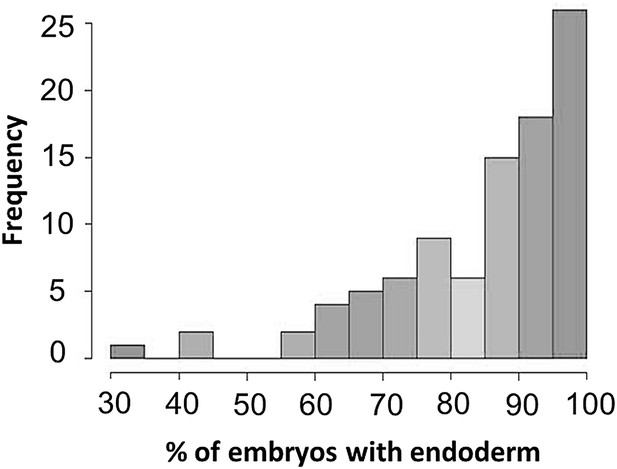
Histogram of mom-2(RNAi) phenotype among the 94 wild isolates.
A beta-distribution is observed (skewed to the right). Shapiro-Wilk normality test (W = 0.8682, p-value=1.207×10−7).
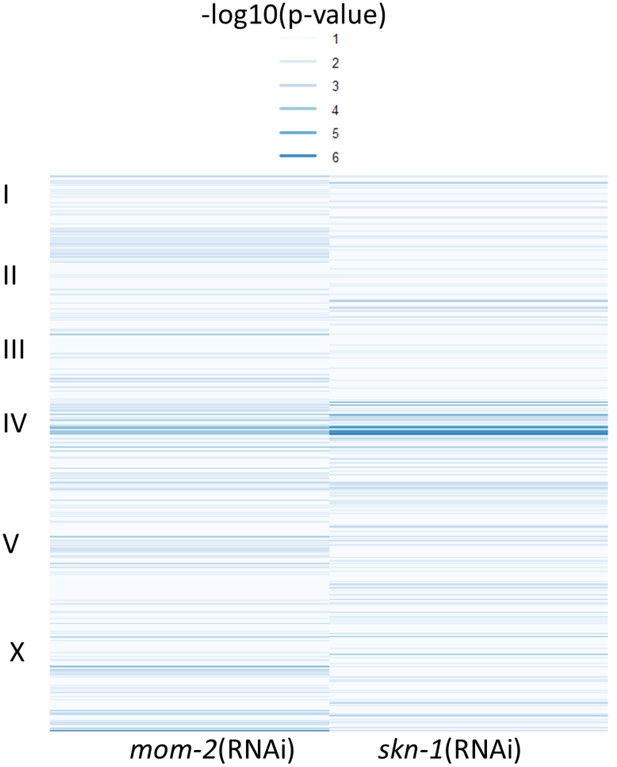
Comparison of EMMA p-values for both mom-2 and skn-1 RNAi phenotypes.
Heatmap of p-values for mom-2(RNAi) (left) and skn-1(RNAi) (right) as calculated in the GWAS analyses (see Figure 5A,B). Strength of association between genotype and endoderm formation phenotypes is represented as –log10(p-value), here depicted as a heatmap (lighter colors – weaker association, darker colors – stronger association). An overlap (indicated by arrowhead) is found in a small region of chromosome IV, but no further correlations are observed. Significant SNPs for skn-1(RNAi) GWAS are shown in Table 1.
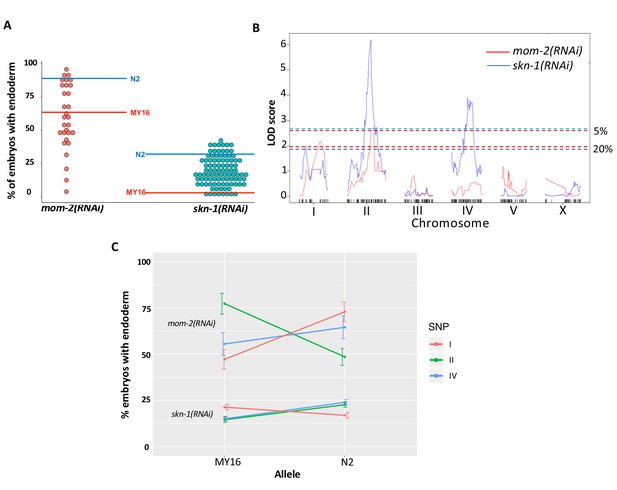
Quantitative genetic analysis of mom-2(RNAi) and skn-1(RNAi) phenotype in Recombinant Inbred Lines (RILs) between N2 and MY16.
(A) mom-2(RNAi) (left) and skn-1(RNAi) (right) phenotype of RILs. The phenotype of the parental strains, MY16 and N2 are shown by red and blue lines, respectively. (B) QTL analyses (interval mapping) of skn-1(RNAi) (blue line) and mom-2(RNAi) (red line) phenotype shown in (A). Genomic regions are shown on the x-axis and LOD score is shown on the y-axis. Significance thresholds for mom-2(RNAi) and skn-1(RNAi) at 5% and 20% linkage represented in red and blue dashed lines, respectively. (C) Effect plots of significant SNPs from mom-2(RNAi) and skn-1(RNAi), indicated by chromosome number and color, showing the direction of the allelic effects. Confidence intervals for the average phenotype in each genotype group are shown.
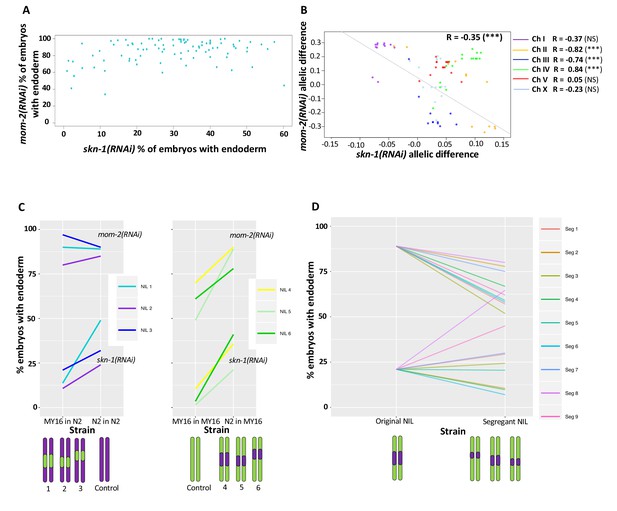
Correlation of skn-1(RNAi) and mom-2(RNAi) allelic differences.
(A) Comparison of skn-1(RNAi) and mom-2(RNAi) phenotype in 94 natural isolates tested. No correlation was found (Spearman correlation R = 0.1844, p-value=0.07). Each dot corresponds to a wild isolate. Y-axis, skn-1(RNAi) phenotype, x-axis, mom-2(RNAi) phenotype. (B) Genome-wide correlation of skn-1(RNAi) and mom-2(RNAi) allelic differences in the N2xMY16 RILs. Each dot represents a SNP. Chromosomes are color-coded with their Pearson’s R values represented (NS = Not Significant, ***=p value<0.001). Regression line in gray. (C) Six different NILs were created for chromosome IV, each of which was compared with a control NIL from the same cross (e.g., MY16 in MY16 as control for N2 in MY16). A schematic of the introgressed regions is represented below the plots. Percentage of skn-1(RNAi) or mom-2(RNAi) embryos with gut is represented. (D) Changes in phenotype for both skn-1(RNAi) and mom-2(RNAi) following recombination in segregant NILs, which a schematic representation of the segregant NILs below the plot.
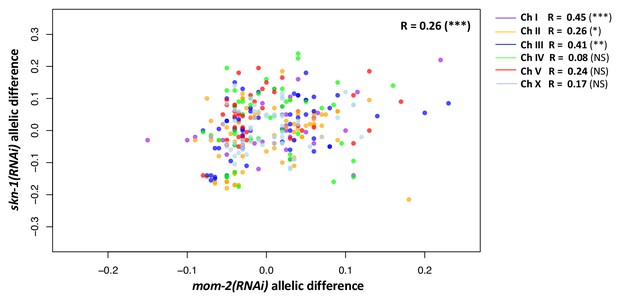
Correlation between the skn-1(RNAi) and mom-2(RNAi) allelic differences.
Each dot represents a SNP, color-coded by chromosome. A small genome-wide correlation is observed, but significant for only three of the chromosomes individually. Represented here is a set of pruned SNPs (N = 321) to cover the whole genome, and corrected for LD (also used for calculations). Z-score is used to calculate median of each allelic group, correcting for outliers. Strength of correlation (Pearson’s R) represented. Significance levels: non-significant, p-value<0.05 (*), p-value<0.01 (**), p-value<0.001 (***).
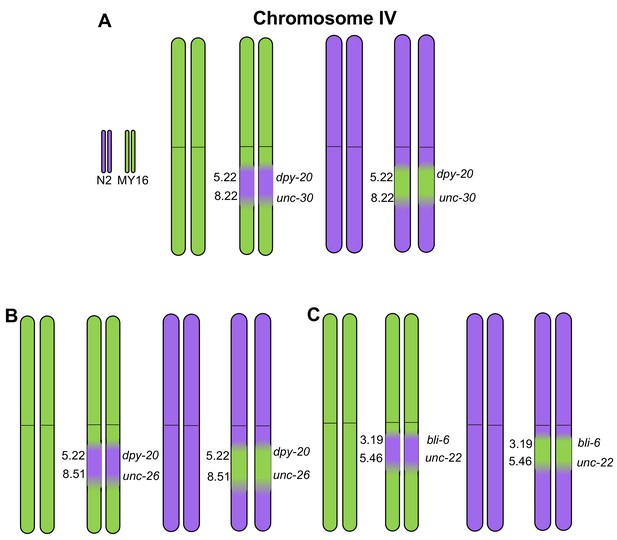
Introgression of N2 and MY16 regions in chromosome IV.
Near-Isogenic Lines (NILs) were built between strains N2 (purple) and MY16 (green), using visual mutations as markers, with their locations in cM indicated in each graph. (A) Introgression of chromosome IV region flanked by dpy-20 and unc-30. (B) Introgression of chromosome IV region flanked by dpy-20 and unc-26. (C) Introgression of chromosome IV region flanked by bli-6 and unc-22.
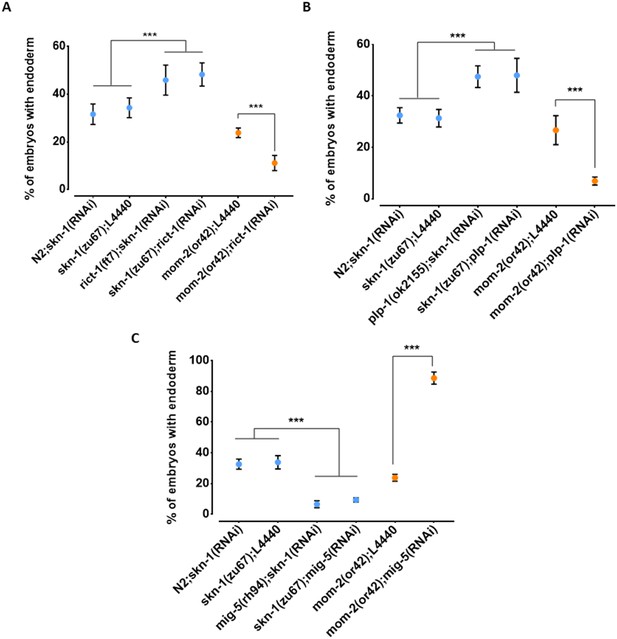
Reciprocal effects of RICT-1, PLP-1, and MIG-5 on skn-1(-) and mom-2(-) phenotypes.
(A, B) Loss of RICT-1 or PLP-1 enhances the mom-2(or42) loss-of-endoderm phenotype and suppresses skn-1(zu67) and skn-1(RNAi) phenotype. (C) Loss of MIG-5 enhances the skn-1(zu67) and skn-1(RNAi) phenotype and suppresses mom-2(or42) phenotype. At least three replicates were performed per experiment and >200 embryos per experiment. Student t-test (*** p-value<0.001). Error bars represent standard deviations.
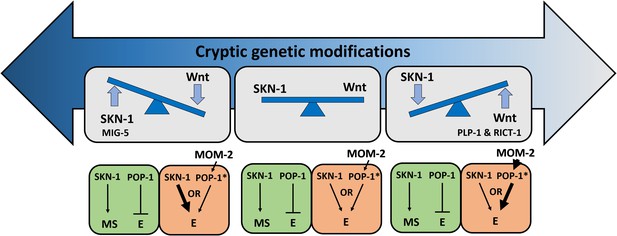
Simplified models for potential cryptic compensatory relationship between the SKN-1 and MOM-2/Wnt regulatory inputs in the endoderm GRN.
Accumulation of cryptic genetic modifications drives rapid rewiring of the GRN, causing broad variation of SKN-1 and MOM-2/Wnt dependence in endoderm (E) specification among C. elegans isotypes. Wnt-signaled POP-1 (indicated by *) acts as an E activator, while unmodified POP-1 in the MS blastomere acts as a repressor of E fate in all C. elegans isotypes. The relative strength of the inputs is indicated by the thickness of the arrow. RICT-1, PLP-1 and MIG-5 reciprocally influence the outcome in the absence of the two inputs.
Tables
Significantly linked SNPs for skn-1(RNAi) GWAS.
https://doi.org/10.7554/eLife.48220.013| SNP | EMMA -log(p) | N2 allele | Variant allele |
|---|---|---|---|
| IV: 5,079,371 bp | 4.957651645 | A | G |
| IV: 5,725,367 bp | 5.211140897 | C | T |
| IV: 5,761,153 bp | 5.211140897 | G | A |
| IV: 5,891,378 bp | 4.252324884 | G | A |
| IV: 5,920,597 bp | 4.720037892 | T | A |
| IV: 5,921,302 bp | 4.720037892 | T | G |
| IV: 5,921,510 bp | 4.252324884 | C | T |
| IV: 6,453,892 bp | 5.142174312 | T | A |
| IV: 6,511,989 bp | 5.142174312 | C | A |
| IV: 6,563,740 bp | 4.678423021 | C | T |
| IV: 7,453,945 bp | 4.652004517 | G | A |
| IV: 7,453,143 bp | 4.181579989 | A | G |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (C. elegans) | Wild isolates; refer to Supplementary file 1 | CGC | ||
| Strain, strain background (C. elegans) | JJ185 | CGC | dpy-13(e184) skn-1(zu67) IV; mDp1 (IV;f). | |
| Strain, strain background (C. elegans) | JR3666 | This study | (elt-2::GFP) X; (ifb-2::CFP) IV; see Figure 1 | |
| Strain, strain background (C. elegans) | EU384 | CGC | dpy-11(e1180) mom-2(or42) V/nT1 [let-?(m435)] (IV;V). | |
| Strain, strain background (C. elegans) | JJ1057 | CGC | pop-1(zu189) dpy-5(e61)/hT1 I; him-5(e1490)/hT1 V. | |
| Strain, strain background (C. elegans) | KQ1366 | CGC | rict-1(ft7) II. | |
| Strain, strain background (C. elegans) | SU351 | CGC | mig-5(rh94)/mIn1 [dpy-10(e128) mIs14] II. | |
| Strain, strain background (C. elegans) | RB1711 | CGC | plp-1(ok2155) IV. | |
| Strain, strain background (C. elegans) | JR3493-JR3590 (refer to Supplementary file 2) | This study | N2xMY16 RILs; see Figure 6 | |
| Strain, strain background (C. elegans) | MT3414 | CGC | dpy-20(e1282) unc-31(e169) unc-26(e205) IV. | |
| Strain, strain background (C. elegans) | DA491 | CGC | dpy-20(e1282) unc-30(e191) IV. | |
| Strain, strain background (C. elegans) | JR2750 | Kontani et al., 2005 | bli-6(Sc16)unc-22(e66)/unc-24(e138)fus-1(w13) dpy-20(e2017)IV | |
| Strain, strain background (C. elegans) | JR3812 (NIL 1) | This study | NIL N2XMY16; see Figure 7 | |
| Strain, strain background (C. elegans) | JR3813 (NIL 2) | This study | NIL N2XMY16; see Figure 7 | |
| Strain, strain background (C. elegans) | JR3814 (NIL 3) | This study | NIL N2XMY16; see Figure 7 | |
| Strain, strain background (C. elegans) | JR3815 (NIL 4) | This study | NIL N2XMY16; see Figure 7 | |
| Strain, strain background (C. elegans) | JR3816 (NIL 5) | This study | NIL N2XMY16; see Figure 7 | |
| Strain, strain background (C. elegans) | JR3817 (NIL 6) | This study | NIL N2XMY16; see Figure 7 | |
| Antibody | MH33 mouse monoclonal | DSHB | RRID:AB_528311 | 1:50 dilution |
| Antibody | AHP418 rabbit polyclonal | Serotec Bio-Rad | RRID:AB_2116715); PMID:28736134 | 1:200 dilution |
| Antibody | ab150116 goat polyclonal | Abcam | RRID:AB_2650601; PMID:31167447 | Goat Anti-Mouse IgG H and L (Alexa Fluor 594) |
| Antibody | ab150077 goat polyclonal | Abcam | RRID:AB_2630356 | Goat Anti-Rabbit IgG H and L (Alexa Fluor 488) |
| Software, algorithm | R v 3.2.3 | The R Foundation | RRID:SCR_001905 | |
| Software, algorithm | PLINK | http://pngu.mgh.harvard.edu/purcell/plink/ | RRID:SCR_001757 |
| position | Ref/Alt | Wild isolates with alt variant (used in this study) |
|---|---|---|
| IV:5,653,761 | C/T | QX1211 |
| IV:5,654,947 | C/T | QX1211 |
| IV:5,655,535 | T/C,A | JU751, MY23, MY16 |
Additional files
-
Supplementary file 1
Wild isolates and their corresponding skn-1(RNAi) and mom-2(RNAi) phenotype, along with isolation and genotype information.
- https://doi.org/10.7554/eLife.48220.020
-
Supplementary file 2
skn-1(RNAi) and mom-2(RNAi) phenotype of N2xMY16 RIL strains.
- https://doi.org/10.7554/eLife.48220.021
-
Supplementary file 3
Genotype data for each of the N2xMY16 RIL strains.
Values of H are Heterozygotes, A is N2 allele, B is MY16 allele, missing allele data represented as “-“.
- https://doi.org/10.7554/eLife.48220.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48220.023





