USP49 potently stabilizes APOBEC3G protein by removing ubiquitin and inhibits HIV-1 replication
Figures
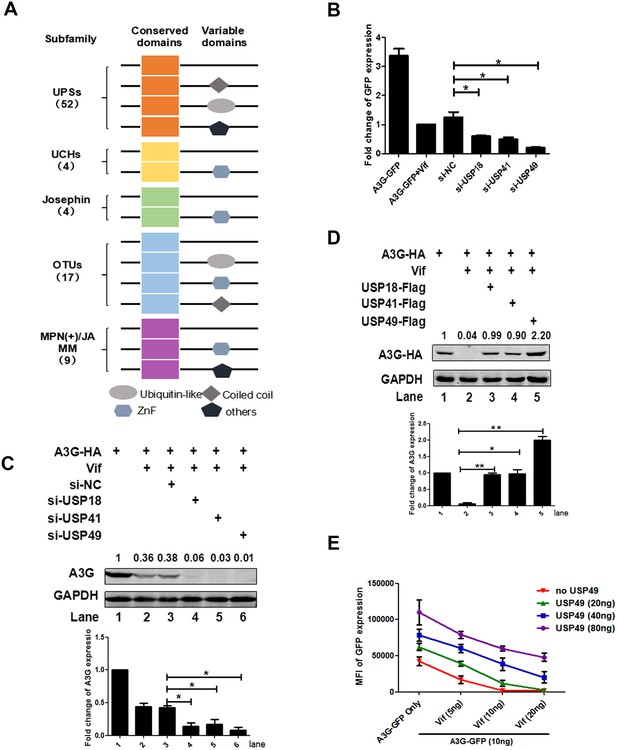
Screening of a DUB-RNAi library identifies that USP 18/41/49 regulates the expression of A3G.
(A) Domain organization of five DUB subfamilies. (B) HEK293T cells were seeded into a 96-well plate with 20,000 cells/well, and then transfected with plasmids expressing A3G-GFP and Vif-HA, as well as siRNAs specific for USP18, USP41, or USP49 respectively. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments.* p<0.05. (C) HEK293T cells were transfected with plasmids expressing A3G-HA and Vif-HA, as well as siRNAs specific for USP18, USP41, or USP49 respectively. After 48 hr, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments.* p<0.05. (D) HEK293T cells were transfected with plasmids expressing A3G-HA, Vif-HA, and one of plasmid expressing USP18-Flag, USP41-Flag, USP49-Flag. After 48 hr, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments.* p<0.05, **p<0.01. (E) HEK293T cells were transfected with indicated amounts of plasmids expressing A3G-GFP, Vif-HA, or USP49-Flag. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments.

The schematic of high-throughput screening.
https://doi.org/10.7554/eLife.48318.003
The distribution of GFP-tagged DUBs and A3G protein.
HEK293T cells were transfected with plasmids expressing USP18-GFP, USP41-GFP, USP49-GFP, and A3G-GFP respectively. After 48 hr, the cells were fixed with 4% paraformaldehyde, and the distribution of DUBs and A3G was detected using a Confocal Microscope.

The expression of endogenous DUBs in primary CD4+ T cells.
(A) Primary CD4+ T cells were isolated from four healthy donors. The mRNA levels of these DUBs were detected by qPCR with their specific primers. Error bars represent the SEM of three independent experiments. (B) The protein level of USP18 in primary CD4+ T cells was detected by western blotting with indicated antibodies. (C) The protein level of USP49 in primary CD4+ T cells was detected by western blotting with indicated antibodies.
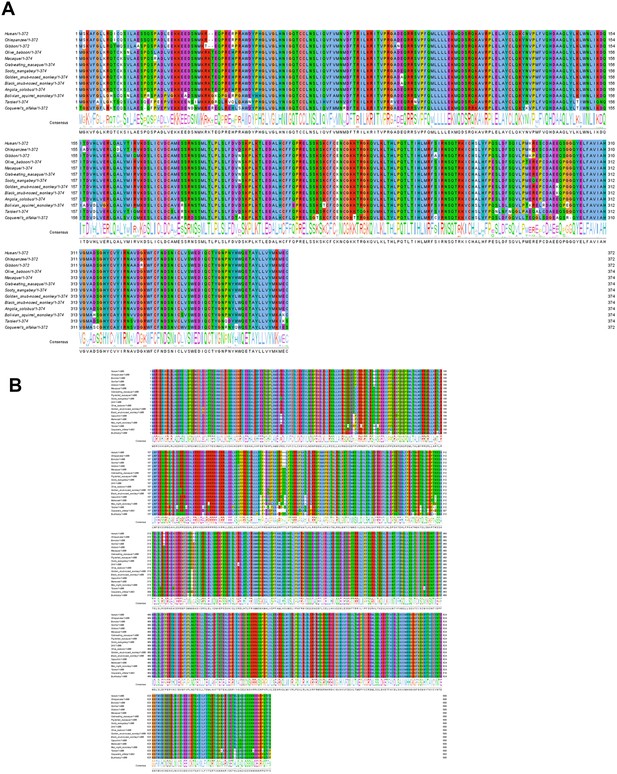
The conservativeness analysis of DUBs among anthropoids.
Primate protein sequences were retrieved from the Ensemble database (http://www.ensembl.org). Orthologues of USP18 (A) and USP49 (B) in other species were confirmed with the comparative genomics from the Ensemble and UCSC Genome databases. Multiple protein sequences alignment was created using the MUSCLE algorithm. Further visualization and conservation analysis with the consensus sequence logos of the multiple sequence alignment were conducted using the Jalview program. The amino acids were colored according to their chemicophysical properties, and the length was shown in the sequence name. Dashes ('-') represent gaps in the sequence.
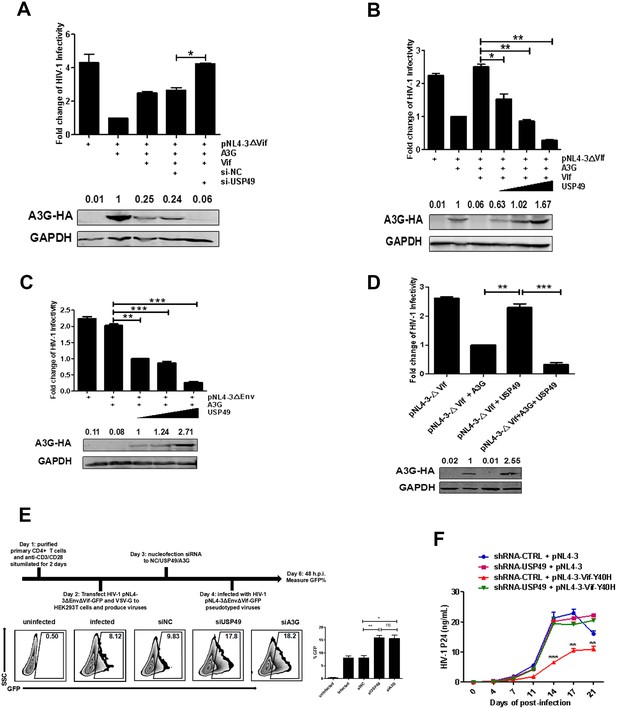
USP49 enhances the inhibitory effect of A3G on the infectivity of HIV-1.
(A) HEK293T cells were first transfected with USP49-specific siRNA. After 12 hr, cells were co-transfected with pcDNA3.1-A3G-HA, pcDNA3.1-Vif-HA, and pNL4-3ΔVif. Culture supernatants were harvested at 72 hr post-transfection and then infected TZM-bl cells. After 48 hr, cells were harvested and the HIV-1 infectivity was detected by luciferase assay. Error bars represent the SEM of three independent experiments. *p<0.05. (B) HEK293T cells were co-transfected with pcDNA3.1-A3G-HA, pNL4-3ΔVif, and plasmids expressing Vif-HA or USP49-Flag respectively. Culture supernatants were harvested at 72 hr post-transfection and then allowed to infect TZM-bl cells. After 48 hr, cells were harvested and the HIV-1 infectivity was detected by luciferase assay. Error bars represent the SEM of three independent experiments. *p<0.05, **p<0.01. (C) HEK293T cells were co-transfected with pcDNA3.1-A3G-HA, pNL4-3ΔEnv, and different amounts of USP49-Flag-expressing plasmid respectively. Culture supernatants were harvested at 72 hr post-transfection and then infected TZM-bl cells. After 48 hr, cells were harvested and the HIV-1 infectivity was detected by luciferase assay. Error bars represent the SEM of three independent experiments. **p<0.01, ***p<0.001. (D) HEK293T cells were transfected with pNL4-3ΔVif, pcDNA3.1-A3G-HA plus pNL4-3ΔVif, or pNL4-3ΔVif plus USP49-Flag plasmids respectively. Culture supernatants were harvested at 72 hr post-transfection and then allowed to infect TZM-bl cells. After 48 hr, cells were harvested and the HIV-1 infectivity was detected by luciferase assay. Error bars represent the SEM of three independent experiments. ***p<0.001. (E) The primary CD4+T cells were stimulated with anti-CD3/28 for 2 days and then nucleofected with indicated siRNAs. After 24 hr, cells were infected with pNL4-3ΔEnvΔVif-GFP pseudotyped viruses. The infectivity were detected by flow cytometr on 48 h.p.i. Representative data were shown and plotted with at least three independent experiments.* p<0.05, **p<0.01. (F) HEK293T cells were transfected with pNL4-3 or Vif-Y40H-mutated-pNL4-3 respectively. Culture supernatants were harvested at 72 hr post-transfection and then allowed to infect shRNA-KD-USP49 primary CD4+ T cells. Cell supernatants were harvested for detecting HIV-1 P24 by ELISA Kit on several time points. Error bars represent the SEM of three independent experiments. The difference of p24 production from HIV-1NL4-3VifY40H infection between shRNA-NC and shRNA-KD-USP49 in primary CD4+ T cells at several time points was statistically analyzed. **p<0.01, ***p<0.001.

Analysis of A3G-induced hypermutation of prot DNA region.
(A) G-to-A mutation loads in proviral DNA from viruses originally produced in Figure 2B (mean + /- SD of 3 independent experiments with a minimum of 10 sequences analyzed per condition). (B) G-to-A mutation loads in proviral DNA from viruses originally produced in Figure 2F (mean + /- SD of 3 independent experiments with a minimum of 10 sequences analyzed per condition).

The A3G protein level was detected by flow cytometry.
The cells from Figrue 2E were fixed with 80% methanol (5 min) and then permeabilized with 0.1% PBS-Tween for 20 min. The cells were then incubated in 1x PBS/10% normal goat serum/0.3M glycine to block non-specific protein-protein interactions followed by the A3G antibody (Abcam, cat#ab75560) for 30 min at 22°C. The secondary antibody used was DyLight 594 goat anti-mouse IgG (H+L) at 1/500 dilution for 30 min at 22°C. Then these cells were detected by flow cytometry and acquisition of >5000 events was performed. Representative data were shown (A) and error bars represent the SEM of three independent experiments (B).
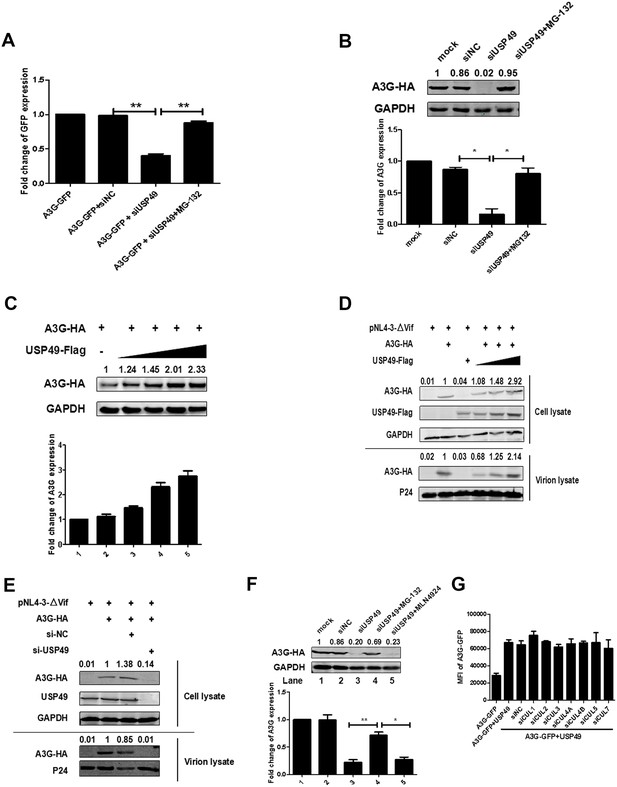
USP49 enhances the expression of A3G even in absence of Vif.
(A) HEK293T cells were transfected with A3G-GFP-expressing plasmid and siRNA specific for USP49. MG132 was treated for 12 hr before harvest. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments. **p<0.01. (B) HEK293T cells were transfected with A3G-HA-expressing plasmid and siRNA specific for USP49. MG132 was treated for 12 hr before harvest. After 48 hr post-transfection, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments.* p<0.05. (C) HEK293T cells were transfected with A3G-HA-expressing plasmid, and different amounts of USP49-Flag-expressing plasmid. After 48 hr, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments. (D) HEK293T cells were transfected with pcDNA3.1-A3G-HA, pcDNA3.1-USP49-Flag, and pNL4-3ΔVif. After 48 hr, cell pellets and supernatants were collected respectively. Cell pellets were lysed and subjected to immunoblotting with anti-HA, anti- Flag, and anti-GAPDH antibodies. Viral particles were collected from filtered supernatants by ultracentrifugation. The pelleted viral particles were lysed and detected by western blotting with anti-HA and anti-p24 antibodies. (E) HEK293T cells were transfected with pcDNA3.1-A3G-HA, pNL4-3, and a USP49-specific siRNA, After 48 hr, cell pellets and supernatants were collected respectively. Cell pellets were lysed and subjected to immunoblotting with anti-HA, anti- Flag, and anti-GAPDH antibodies. Viral particles were collected from filtered supernatants by ultracentrifugation. The pelleted viral particles were lysed and detected by western blotting with anti-HA and anti-p24 antibodies. (F) HEK293T cells were transfected with pcDNA3.1-A3G-HA and a USP49-specific siRNA. MG132 (10 uM) or MLN4924 (20 uM) was treated for 12 hr before harvest. After 48 hr post-transfection, cells were lysed and Western blot was performed with the indicated antibodies. Representative data were shown and plotted with at least three independent experiments.* p<0.05, **p<0.01. (G) HEK293T cells were transfected with pcDNA3.1-A3G-GFP, pcDNA3.1-USP49-FLAG and indicated siRNAs. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments.
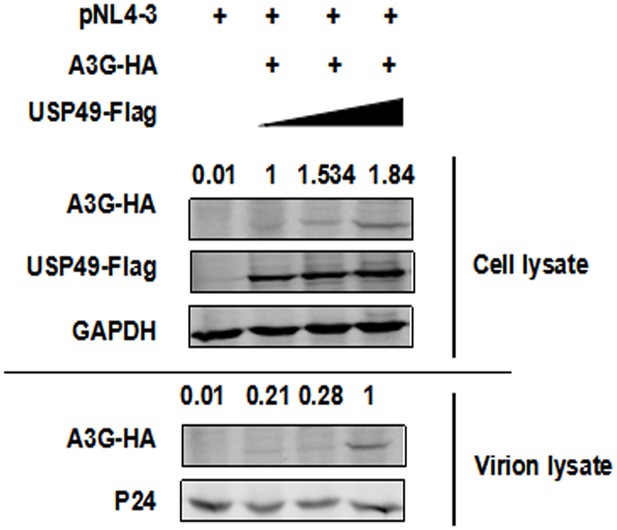
A3G can be packaged into HIV-1 virions with the help of USP49.
HEK293T cells were transfected with pcDNA3.1-A3G-HA, pcDNA3.1-USP49-Flag, and pNL4-3. After 48 hr, cell pellets and supernatants were collected respectively. Cell pellets were lysed and subjected to immunoblotting with anti-HA, anti- Flag, and anti-GAPDH antibodies. Viral particles were collected from filtered supernatants by ultracentrifugation. The pelleted viral particles were lysed and detected by western blotting with anti-HA and anti-p24 antibodies.
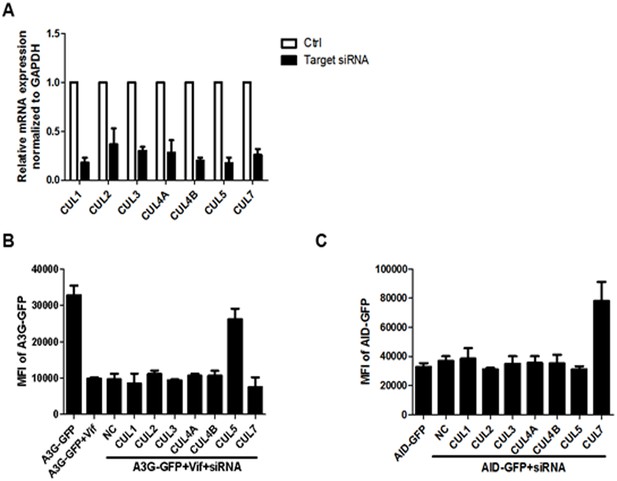
Detection of the knockdown efficiency of various siRNAs.
(A) HEK293T cells were transfected with indicated siRNAs. After 48 hr, the total RNA was extracted with TRIZOL and the mRNA expression of each gene was detected by qRT-PCR. Error bars represent the SEM of three independent experiments. (B) HEK293T cells were transfected with pcDNA3.1-A3G-GFP, pcDNA3.1-Vif, and indicated siRNAs. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments. (C) HEK293T cells were transfected with pcDNA3.1-AID-GFP and the indicated siRNAs. The GFP expression was detected with a PE Envision at 48 hr post-transfection. Error bars represent the SEM of three independent experiments.
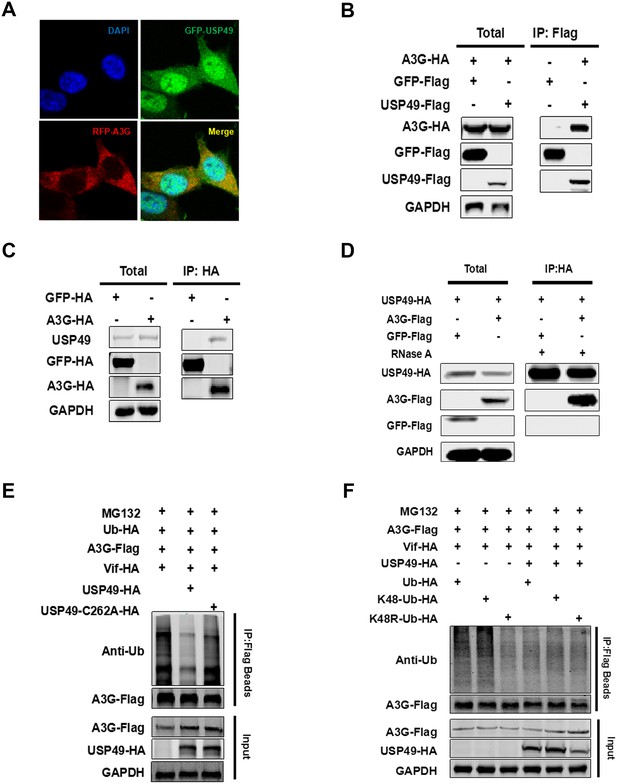
USP49 directly interacts with A3G and deubiquitinates the K48-linked ubiquitination of A3G.
(A) HEK293T cells were co-transfected with plasmids expressing USP49-GFP or A3G-RFP. After 48 hr, the cells were fixed with 4% paraformaldehyde, and localization of USP49 and A3G was detected using confocal microscopy. (B) HEK293T cells were co-transfected with plasmids expressing Flag-USP49 or HA-A3G, or the indicated vectors. The cell lysates were immunoprecipitated using Flag beads and analyzed by immunoblotting with the indicated antibodies. (C) Hela cells were co-transfected with plasmids expressing HA-GFP or HA-A3G. The cell lysates were immunoprecipitated using HA beads and analyzed by immunoblotting with the indicated antibodies. (D) HEK293T cells were co-transfected with plasmids expressing HA-USP49 and Flag-A3G. The cell lysates were immunoprecipitated using HA beads and then treated with RNase A (20 ug/mL) for 1 hr. After that, samples were analyzed by immunoblotting with the indicated antibodies. (E) HEK293T cells were co-transfected with plasmids expressing USP49-HA, Vif-HA, Ub-HA, or A3G-Flag. Cells were treated with 10 uM MG132 for 24 hr before harvest. After 48 hr post-transfection, the cells were subjected to the denaturing immunoprecipitation using an anti-Flag beads followed by immunoblot analysis using the indicated antibodies. (F) HEK293T cells were transfected with the indicated plasmids expressing HA-ubiquitin, Flag-A3G, HA-Vif, or HA-USP49. After 24 hr, the cells were treated with 10 μM MG132 for 24 hr; the indicated types of ubiquitination were detected and quantified by the denaturing immunoprecipitation and western blotting.
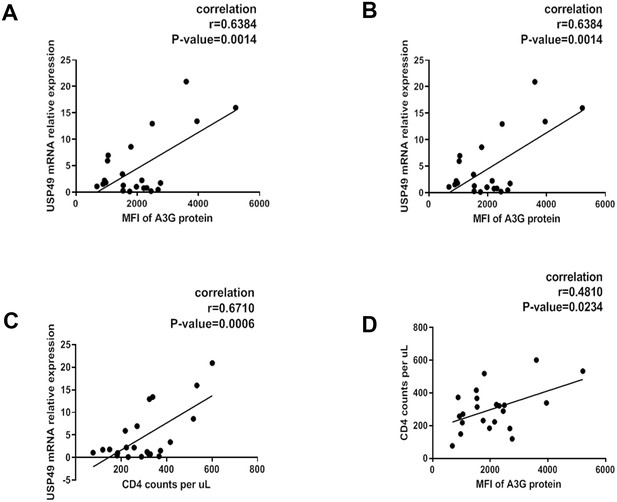
Associations of plasma HIV-1 RNA levels, CD4+ T cell counts and USP49 mRNA expression levels in the newly-diagnosed HIV-1-infected individuals.
(A) Correlation between the expression of A3G protein level and the USP49 mRNA level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals (n = 21). Pearson correlation coefficient and p value are listed. (B) Correlation between the count of CD4+T cells and the USP49 mRNA level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals. Pearson correlation coefficient and p value are listed. (C) Correlation between the count of CD4+T cells and the A3G protein level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals. Pearson correlation coefficient and p value are listed. (D) Correlation between the plasma HIV-1 RNA levels and the USP49 mRNA level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals. Pearson correlation coefficient and p value are listed.
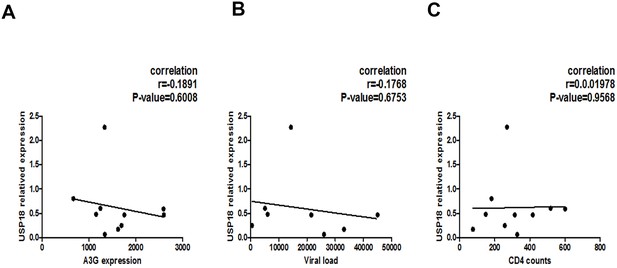
Associations of plasma HIV-1 RNA levels, CD4+ T cell counts, and USP18 mRNA expression levels in the newly-diagnosed HIV-1-infected individuals.
(A) Correlation between the expression of A3G and the USP18 mRNA level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals (n = 10). Pearson correlation coefficient and p value are listed. (B) Correlation between the count of CD4+T cells and the USP18 mRNA level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals. Pearson correlation coefficient and p value are listed. (C) Correlation between the plasma HIV-1 RNA levels and the USP18 mRNA level in the CD4+T cells isolated from the newly-diagnosed HIV-1-infected individuals. Pearson correlation coefficient and p value are listed.
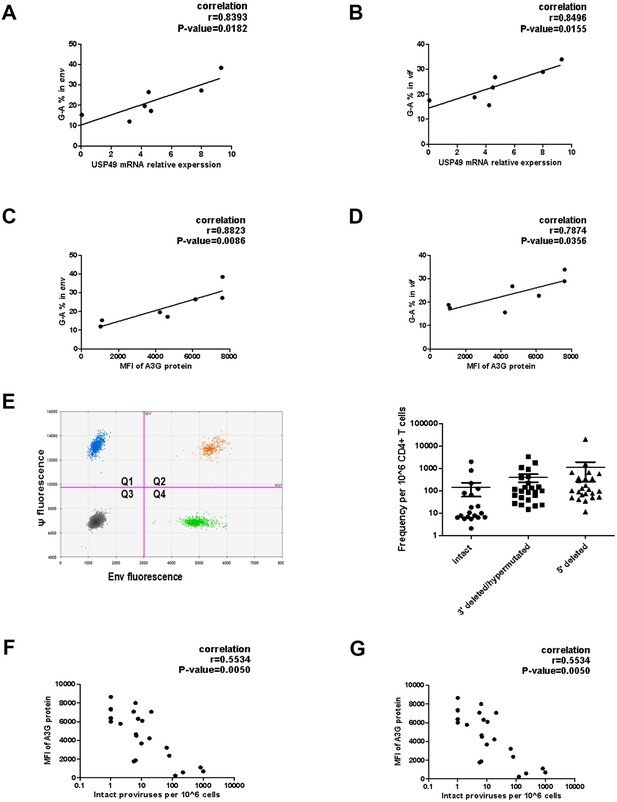
Associations of defective proviruses and USP49 mRNA expression levels in the CD4+ T cells isolated from HIV-1-infected individuals receiving suppressive cART.
(A–B) The percentage of G-to-A in env and vif population sequences correlates with USP49 mRNA expression level in the CD4+ T cells from HIV-1-infected individuals (n = 7). Pearson correlation coefficient and p value are listed. (C–D) The percentage of G-to-A in env and vif population sequences correlates with A3G protein level in the CD4+ T cells from HIV-1-infected individuals (n = 7). Pearson correlation coefficient and p value are listed. (E) Representative IPDA results from a patient’s CD4+ T cell sample. Boxed areas are expanded to show individual positive droplets (Left). IDPA results on CD4+ T cells from HIV-1-infected individuals (n = 24) with plasma HIV-1 RNA below the limit of detection (right). (F) Correlation between the intact proviruses and the A3G protein level in the CD4+T cells isolated from the indicated clinical HIV-1 patients. Pearson correlation coefficient and p value are listed. (G) Correlation between the intact proviruses and the USP49 mRNA level in the CD4+T cells isolated from the indicated clinical HIV-1 patients. Pearson correlation coefficient and p value are listed.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | E. coli DH5α: F-, φ 80dlacZ ΔM15, Δ(lacZYA -argF) U169, deoR, recA1, endA1, hsdR17 (rK-, mK+), phoA, supE44, λ-, thi −1, gyrA96, relA1 | Takara | Cat#9057 | |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216; RRID: CVCL_0063 | female |
| Cell line (Homo sapiens) | Hela | ATCC | CCL-2; RRID: CVCL_0030 | female |
| Cell line (Homo sapiens) | TZM-bl | NIH AIDS Reagent Program | Cat#8129 | female |
| Biological sample (Homo sapiens) | Blood samples from healthy individuals | Guangzhou Blood Center, Guangzhou | http://www.gzbc.org/ | |
| Biological sample (Homo sapiens) | Blood samples from HIV-1-infected individuals | Department of Infectious Diseases, Guangzhou 8th People’s Hospital, Guangzhou | http://gz8h.com.cn/ | |
| Antibody | Mouse Monoclonal Anti-HA-Tag Antibody | MBL | Cat#M180-3 | Dilution 1:1000 |
| Antibody | Rabbit Anti-DDDDK Tag Polyclonal Antibody, Unconjugated | MBL | Cat#PM020 | Dilution 1:1000 |
| Antibody | Rabbit Polyclonal Anti-GAPDH Antibody | Proteintech | Cat#10494–1-AP | Dilution 1:1000 |
| Antibody | beta Actin Mouse McAb | Proteintech | Cat#66009–1-Ig | Dilution 1:1000 |
| Antibody | USP49 Rabbit Polyclonal antibody | Proteintech | Cat#18066–1-AP | Dilution 1:500 |
| Antibody | USP18 (D4E7) Rabbit mAb | Cell Signaling Technology (CST) | Cat#4813 | Dilution 1:1000 |
| Antibody | Anti-APOBEC3G/A3G antibody | Abcam | Cat#ab75560 | Dilution 1:200 |
| Antibody | GFP (D5.1) XP Rabbit mAb | Cell Signaling Technology (CST) | Cat#2956 | Dilution 1:1000 |
| Antibody | Ubiquitin Rabbit Polyclonal antibody | Proteintech | Cat#10201–2-AP | Dilution 1:1000 |
| Antibody | IRDye 680RD Goat anti-Mouse IgG (H + L), 0.5 mg Antibody | LI-COR Biosciences | Cat#926–68070 | Dilution 1:10000 |
| Antibody | IRDye 800CW Goat Anti-Rabbit IgG, Conjugated Antibody | LI-COR Biosciences | Cat#926–32211 | Dilution 1:10000 |
| Antibody | Goat Anti-Mouse IgG H and L (DyLight 488) preadsorbed | Abcam | Cat#ab96879 | Dilution 1:500 |
| Antibody | Goat Anti-Mouse IgG H and L (DyLight 594) preadsorbed | Abcam | Cat#ab96881 | Dilution 1:500 |
| Antibody | EZviewTM Red Anti-HA Affinity Gel | Sigma-Aldrich | Cat# A2220-10ML | 30 ul/sample |
| Antibody | ANTI-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat# E6779-1ML | 30 ul/sample |
| Recombinant DNA reagent | VSV-G glycoprotein-expression vector | PMID: 9306402 | Addgene Plasmid #12259 | Dr. Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) |
| Recombinant DNA reagent | Lentiviral packaging construct pCMVΔR8.2 | PMID: 9306402 | Addgene Plasmid #12263 | Dr. Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) |
| Sequence-based reagent | siRNA Library | RiboBio | http://www.ribobio.com/ | |
| Chemical compound, drug | TRIzolTM Reagent | ThermoFisher | Cat#15596018 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat#T8787-50ML | |
| Chemical compound, drug | Penicillin-Streptomycin,Liquid | ThermoFisher | Cat#15140122 | |
| Commercial assay or kit | BD IMag Human CD4+ T Lymphocyte Enrichment Set-DM | BD Biosciences | Cat#557939 | |
| Commercial assay or kit | HIV-1 p24 ELISA Kit | Abcam | Cat#ab218268 | |
| Software, algorithm | Prism 5 | GraphPad | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | FlowJo V10 | Tree Star | https://www.flowjo.com/ | |
| Software, algorithm | Odyssey CLX Imager | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/odyssey/ | |
| Software, algorithm | Image Studio Lite Ver 4.0 | LI-COR Biosciences | https://www.licor.com/bio/products/software/image_studio_lite/ | |
| Software, algorithm | Hypermut | https://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html |
Additional files
-
Supplementary file 1
The sequences of primers and probes.
- https://doi.org/10.7554/eLife.48318.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48318.018





