Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence
Figures
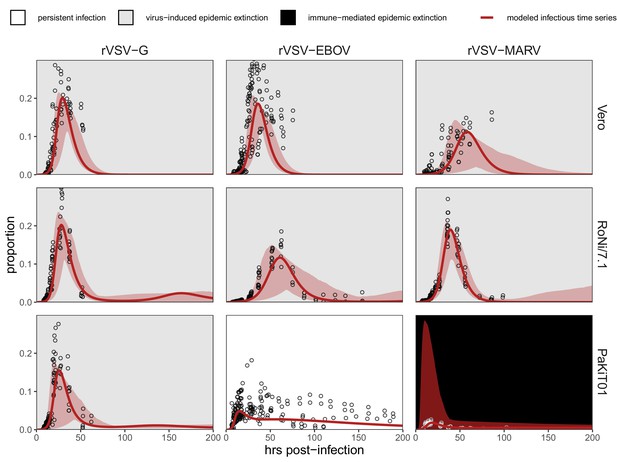
Fitted time series of infectious cell proportions from mean field model for rVSV-G, rVSV-EBOV, and rVSV-MARV infections (columns) on Vero, RoNi/7.1, and PaKiT01 cell lines (rows) at MOI = 0.001.
Results are shown for the best fit immune absent model on Vero cells, induced immunity model on RoNi/7.1 cells, and constitutive (for rVSV-VSVG and rVSV-EBOV) and induced (for rVSV-MARV) immunity models on PaKiT01 cells. Raw data across all trials are shown as open circles (statistical smoothers from each trial used for fitting are available in Figure 1—figure supplements 2–3). Model output is shown as a solid crimson line (95% confidence intervals by standard error = red shading). Panel background corresponds to empirical outcome of the average stochastic cell culture trial (persistent infection = white; virus-induced epidemic extinction = gray; immune-mediated epidemic extinction = black). Parameter values are listed in Table 1 and Supplementary file 4. Results for absent/induced/constitutive fitted models across all cell lines are shown in Figure 1—figure supplement 4 (MOI = 0.001) and Figure 1—figure supplement 5 (MOI = 0.0001).
Cell culture models of viral propagation.
(A), (B), and (C) show raw, original images of rVSV-EBOV propagation across Vero cell lines at, respectively, 17, 21, and 28 hr post-infection (timesteps 2, 3, and five from trial Ver6_B1). (D), (E), and (F) show corresponding, binary images processed in the R package, EBImage. Cells expressing viral eGFP are depicted in white and uninfected/dead cells in black.
Time series data to which mean field mechanistic models were fit, across rVSV-G (left), rVSV-EBOV (middle), and rVSV-MARV (right) infections on Vero, RoNi/7.1, and PaKiT01 cell lines, at MOI = 0.001.
Open circles show raw data across all trials, while red, dashed line gives the statistical mean of each trials, established from GAM model incorporating random effects per trial. Results for MOI = 0.0001 are shown in Figure 1—figure supplement 3.
Time series data to which mean field mechanistic models were fit, across rVSV-G (left), rVSV-EBOV (middle), and rVSV-MARV (right) infections on Vero, RoNi/7.1, and PaKiT01 cell lines, at MOI = 0.0001.
Open circles show raw data across all trials, while red, dashed line gives the statistical mean of each trials, established from GAM model incorporating random effects per trial. Results for MOI = 0.001 are shown in Figure 1—figure supplement 2.
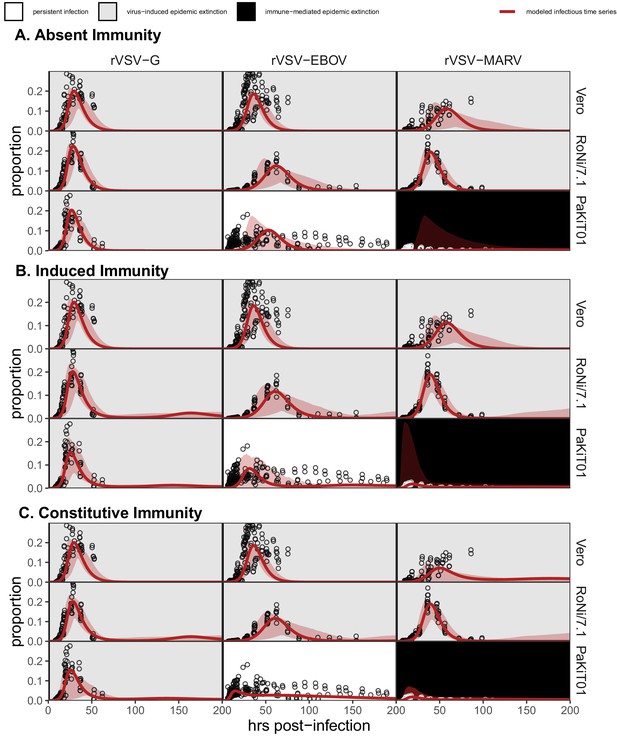
Figure replicates Figure 1 (main text) but includes all output across mean field model fits assuming (A) absent immunity, (B) induced immunity, and (C) constitutive immunity.
Figure shows fitted time series of infectious cell proportions for rVSV-G, rVSV-EBOV, and rVSV-MARV infections (columns) on Vero, RoNi/7.1, and PaKiT01 cell lines (rows) at MOI = 0.001. Raw data across all trials are shown as open circles and model output as the solid crimson line (95% confidence intervals by standard error = red shading). Panel background corresponds to empirical outcome of the average stochastic cell culture trial (persistent infection = white; virus-induced epidemic extinction = gray; immune-mediated epidemic extinction = black).
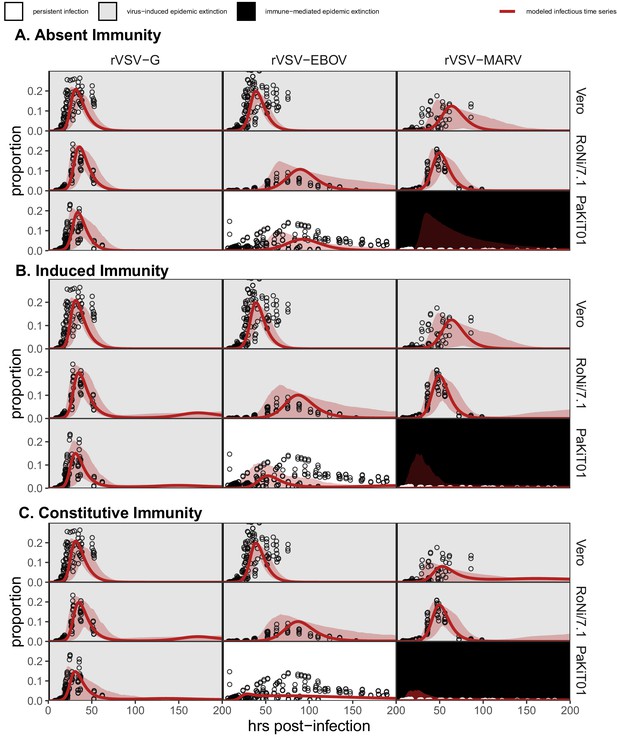
Figure replicates Figure 1—figure supplement 4 exactly but shows model fits and data for all cell-virus combinations at MOI = 0.0001.
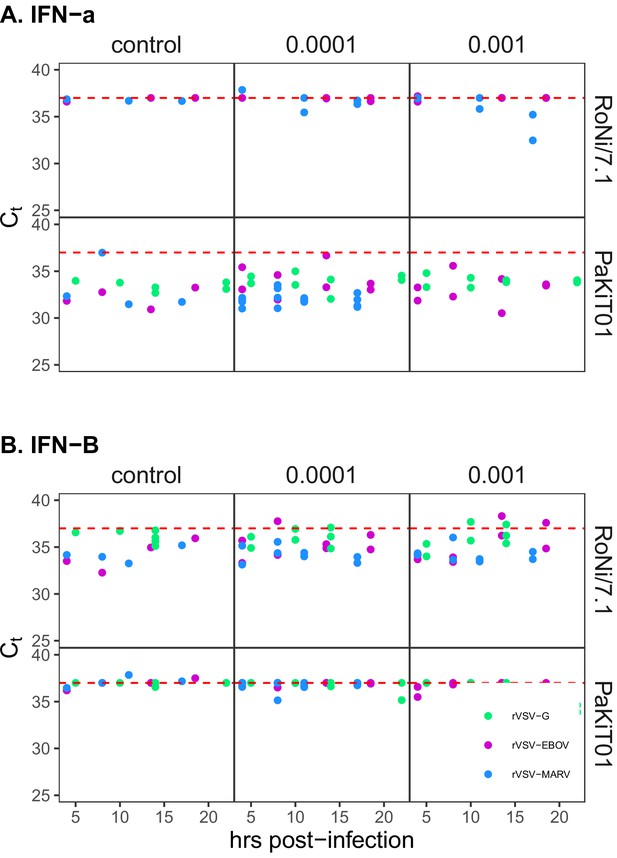
IFN gene expression in bat cells at baseline and upon viral stimulation.
(A) IFN-α and (B) IFN-β gene expression profiles from qPCR for rVSV infections on RoNi/7.1 and PaKiT01 cell lines. Panels show δ-Ct (raw Ct of IFN gene assay subtracted from raw Ct of β-Actin housekeeping gene assay) across a time series for mock (left), MOI = 0.0001 (middle) and MOI = 0.001 (right) infections across a time series. Viruses are represented by color (rVSV-G = green, rVSV-EBOV = magenta, rVSV-MARV = blue). The red dashed line at δ-Ct = 37 corresponds to no expression; higher expression is indicated at lower values for δ-Ct. qPCR was carried out using primers summarized in Supplementary file 6.
Curve fits to control data for standard birth (b = .025) and natural mortality ( hours for, respectively, Vero, RoNi/7.1, and PaKiT01 cell lines) rates across all three cell lines.
Raw data from multiple trials are shown as open circles, statistical means as dashed black lines, with the output from the mean field model, using the fixed birth rate and estimated mortality rate, in solid green.
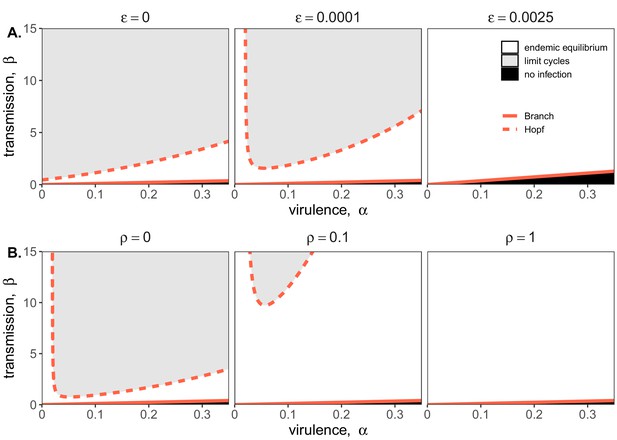
Two parameter bifurcations of the mean field model, showing variation in the transmission rate, β, against variation in the pathogen-induced mortality rate, α, under diverse immune assumptions.
Panel (A) depicts dynamics under variably constitutive immunity, ranging from absent (left: ) to high (right: ). In all panel (A) plots, the rate of induced immune antiviral acquisition (ρ) was fixed at 0.01. Panel (B) depicts dynamics under variably induced immunity, ranging from absent (left: ρ=0) to high (right: ρ=1). In all panel (B) plots, the rate of constitutive antiviral acquisition () was fixed at 0.0001 Branch point curves are represented as solid lines and Hopf curves as dashed lines. White space indicates endemic equilibrium (persistence), gray space indicates limit cycles, and black space indicates no infection (extinction). Other parameter values for equilibrium analysis were fixed at: b = .025, μ = .001, σ = 1/6, c = 0. Special points from bifurcations analyses are listed in Supplementary file 3.
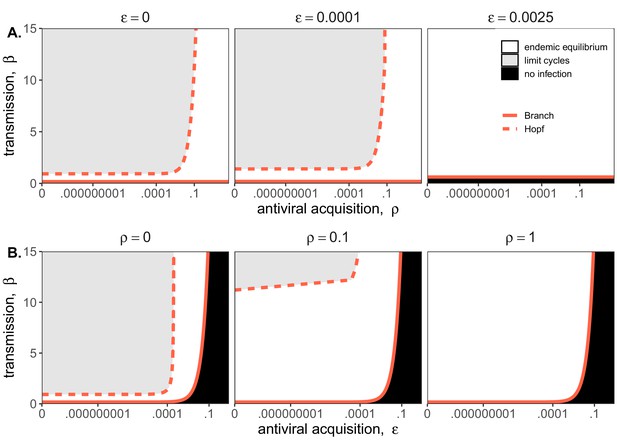
Two parameter bifurcations of the mean field model, showing variation in the transmission rate, β, against variation in: (A) the induced immunity rate of antiviral acquisition (ρ) and (B) the constitutive immunity rate of antiviral acquisition ().
Panels show variation in the extent of immunity, from absent (left) to high (right). Branch point curves are represented as solid lines and Hopf curves as dashed lines. White space indicates endemic equilibrium (persistence), gray space indicates limit cycling, and black space indicates no infection (extinction). Other parameter values for equilibrium analysis were fixed at: b = .025, μ = .001, σ = 1/6, α = 1/6, c = 0. Special points from bifurcations analyses are listed in Supplementary file 3.
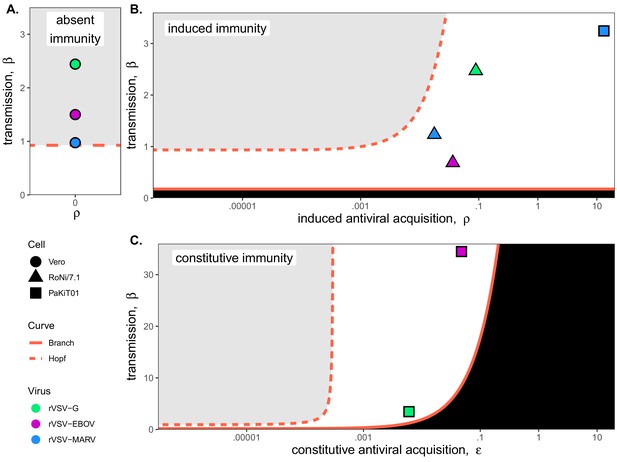
Best fit parameter estimates for β and ρ or from mean-field model fits to MOI=0.001 time series data, atop (A,B) β – ρ and (C) β – bifurcation.
Fits and bifurcations are grouped by immune phenotype: (A) absent; (B) induced; (C) constitutive immunity, with cell lines differentiated by shape (Vero=circles; RoNi/7.1 = triangles; PaKiT01=squares) and viral infections by color (rVSV-G = green, rVSV-EBOV = magenta, rVSV-MARV = blue). Note that y-axis values are ten-fold higher in panel (C). Branch point curves (solid lines) and Hopf curves (dashed lines) are reproduced from Figure 3. White space indicates endemic equilibrium (pathogen persistence), gray space indicates limit cycling (virus-induced epidemic extinction), and black space indicates no infection (immune-mediated pathogen extinction). In panel (A) and (B), is fixed at 0; in panel (C), ρ is fixed at 5x10−8 for bifurcation curves and estimated at 4x10−8 and 8x10−8 for rVSV-EBOV and rVSV-G parameter points, respectively. Other parameter values were fixed at: b = .025, μ = 0.001, σ = 1/6, α = 1/6, and c = 0 across all panels. Raw fitted values and corresponding 95% confidence intervals for β, ρ, and , background parameter values, and AIC recovered from model fit, are reported in Supplementary file 4. Parameter fits at MOI=0.0001 are visualized in Figure 4—figure supplement 1.
Best fit parameter estimates for β and ρ or from mean-field model fits to MOI=0.0001 time series data, atop (A,B) β – ρ and (C) β – bifurcation.
Fits and bifurcations are grouped by immune phenotype: (A) absent; (B) induced; (C) constitutive immunity, with cell lines differentiated by shape (Vero=circles; RoNi/7.1 = triangles; PaKiT01=squares) and viral infections by color (rVSV-G = green, rVSV-EBOV = magenta, rVSV-MARV = blue). Note that y-axis values are ten-fold higher in panel (C). Branch point curves (solid lines) and Hopf curves (dashed lines) are reproduced from Figure 3 (main text). White space indicates endemic equilibrium (pathogen persistence), gray space indicates limit cycling (virus-induced epidemic extinction), and black space indicates no infection (immune-mediated pathogen extinction). In panel (A) and (B), is fixed at 0; in panel (C), is fixed at 5x10−8 for bifurcation curves and estimated at 4x10−8 and 8x10−8 for rVSV-EBOV and rVSV-G parameter points, respectively. To construct bifurcation curves, other parameter values were fixed at: b = 0.025, µ = 0.001, , and c = 0 across all panels. Raw fitted values and corresponding 95% confidence intervals for β, ρ, and , background parameter values, and AIC recovered from model fit, are reported in Supplementary file 4. Parameter fits at MOI=0.0001 are visualized in Figure 4 of the main text.
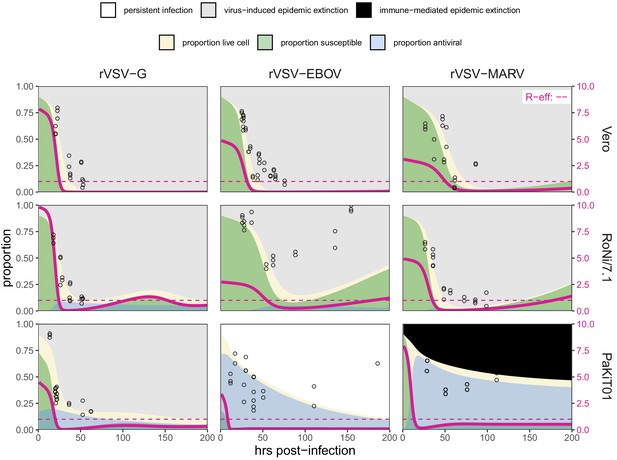
Fitted time series of susceptible (green shading) and antiviral (blue shading) cell proportions from the mean field model for rVSV-G, rVSV-EBOV, and rVSV-MARV infections (columns) on Vero, RoNi/7.1, and PaKiT01 cell lines (rows) at MOI = 0.001.
Results are shown for the best fit immune absent model on Vero cells, induced immunity model on RoNi/7.1 cells and constitutive (rVSV-G and rVSV-EBOV) and induced (rVSV-MARV) immune models on PaKiT01 cells. Combined live, uninfectious cell populations (S + A + E) are shown in tan shading, with raw live, uninfectious cell data from Hoechst stains visualized as open circles. The right-hand y-axis corresponds to R-effective (pink solid line) across each time series; R-effective = 1 is a pink dashed, horizontal line. Panel background corresponds to empirical outcome of the average stochastic cell culture trial (persistent infection = white; virus-induced epidemic extinction = gray; immune-mediated epidemic extinction = black). Parameter values are listed in Supplementary file 4 and results for absent/induced/constitutive fitted models across all cell lines in Figure 5—figure supplement 1 (MOI = 0.001) and Figure 5—figure supplement 2 (MOI = 0.0001).
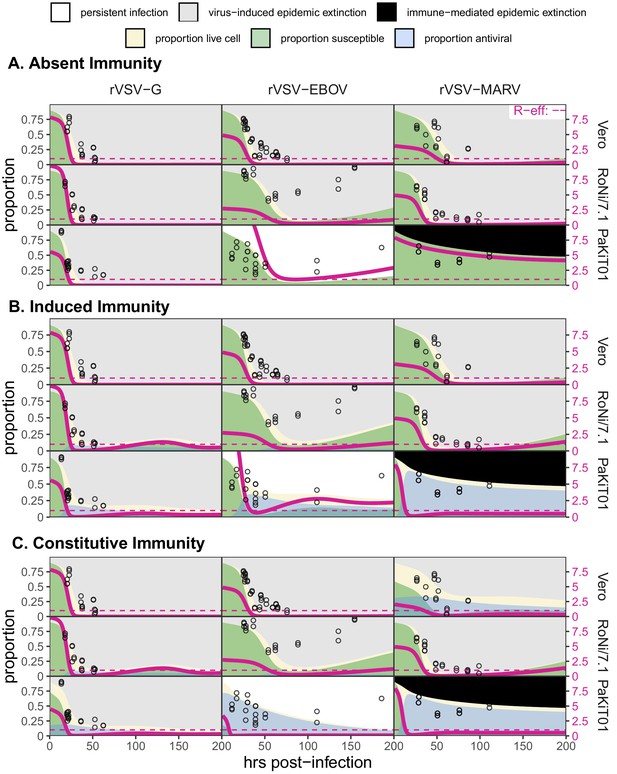
Figure replicates Figure 5 (main text) but includes all output across mean field model fits assuming (A) absent immunity, (B) induced immunity, and (C) constitutive immunity.
Figure shows fitted time series of susceptible (green shading) and antiviral (blue shading) cell proportions from the mean field model for rVSV-G, rVSV-EBOV, and rVSV-MARV infections (columns) on Vero, RoNi/7.1, and PaKiT01 cell lines (rows) at MOI = 0.001. Combined live, uninfectious cell populations (S + A + E, summed across the time series) is shown in tan shading, with raw live, uninfectious cell data from Hoechst stains of terminal time series visualized as open circles. The right-hand y-axis corresponds to R-effective (pink solid line) across each time series; R-effective = 1 is given as a pink dashed, horizontal line. Panel background corresponds to empirical outcome of the average stochastic cell culture trial (persistent infection = white; virus-induced epidemic extinction = gray; immune-mediated epidemic extinction = black).
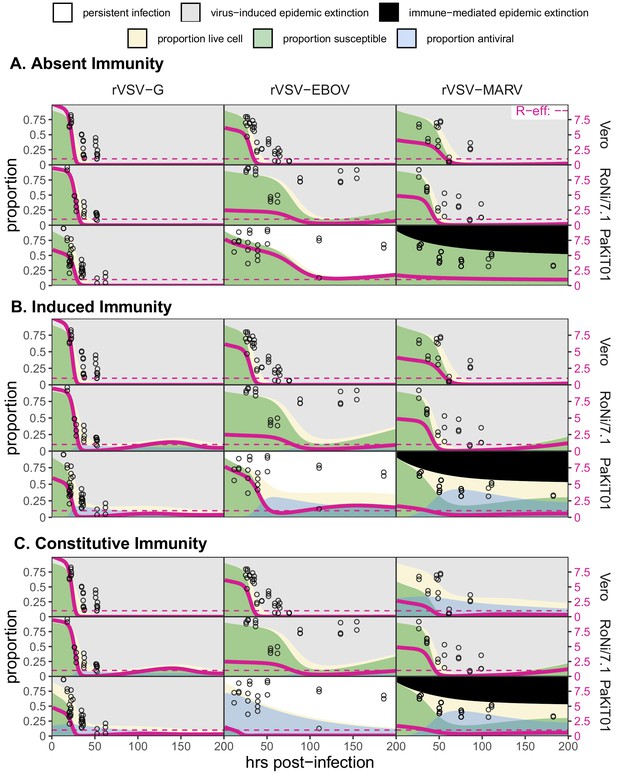
Figure replicates Figure 5—figure supplement 1 exactly but shows model fits and data for all cell-virus combinations at MOI = 0.0001.
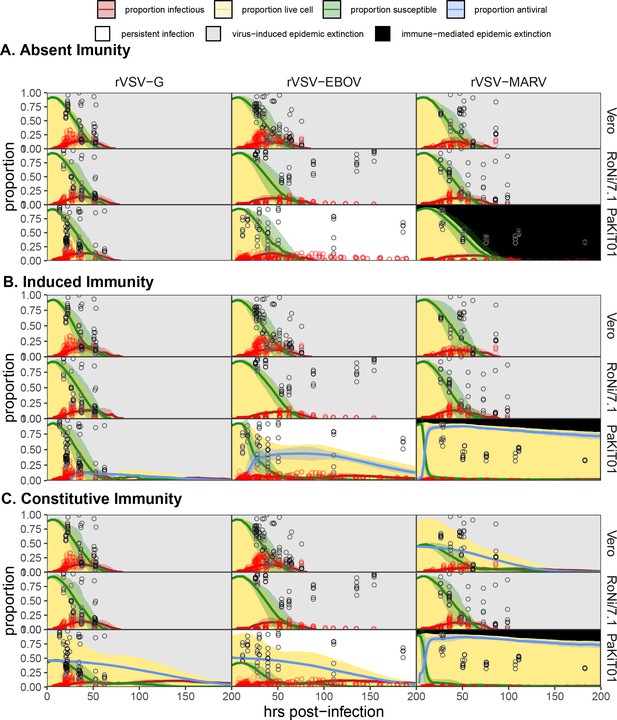
Spatial model state variable outputs, fit to MOI = 0.001 data only, for all 27 unique cell line - virus - immune assumption combinations: (A) absent immunity, (B) induced immunity, and (C) constitutive immunity.
Values for ρ and were fixed at equivalent values to those optimized in mean field trials and β fixed at ten times the value estimated under mean field conditions. Figure shows mean output from 10 runs of the spatial stochastic model, on a 10,000 cell lattice for MOI = 0.001 infections of rVSV-G, rVSV-EBOV, and rVSV-MARV (columns) on Vero, RoNi/7.1, and PaKiT01 (rows) cell lines. Mean state variable outputs are plotted as colored lines with 95% confidence intervals by standard error shown in corresponding shading (infectious = red; susceptible = green; antiviral = blue). Raw infectious cell data across all time trials are plotted as open red circles, with the Hoechst-stained live cell population as open black circles. Modeled live, uninfectious cell populations (S+A+E) are shown in tan shading in the background. Panel background shading corresponds to the mean spatial model outcome for each cell line – virus combination (persistent infection = white; virus-induced epidemic extinction = gray; immune-mediated epidemic extinction = black). All parameter values are reported in Supplementary file 4.
Videos
Two hundred hour time series of spatial stochastic model for rVSV-EBOV infection on 10,000 cell grid for PaKiT01, assuming conditions of absent immunity: (A) spatial spread of infection, (B) time series of state variables.
Parameter values are listed in Supplementary file 4.
Two hundred hour time series of spatial stochastic model for rVSV-EBOV infection on 10,000 cell grid for PaKiT01, assuming conditions of induced immunity: (A) spatial spread of infection, (B) time series of state variables.
Parameter values are listed in Supplementary file 4.
Two hundred hour time series of spatial stochastic model for rVSV-EBOV infection on 10,000 cell grid for PaKiT01, assuming conditions of constitutive immunity: (A) spatial spread of infection, (B) time series of state variables.
Parameter values are listed in Supplementary file 4.
Tables
Optimized parameters from best fit deterministic model and spatial approximation at MOI = 0.001
| Cell line | Virus | Immune assumption | AIC reduction from next-best model | Antiviral rate | [lci – uci] * | ρ [lci – uci] * | β [lci – uci] * | Mean field R0 | Spatial β |
|---|---|---|---|---|---|---|---|---|---|
| Vero | rVSV-G | Absent | 2 | 0 | 0 [0–0] | 0 [0–0] | 2.44 [1.52–3.36] | 8.73 | 24.418 |
| rVSV-EBOV | Absent | 2 | 0 | 0 [0–0] | 0 [0–0] | 1.5 [1.06–1.94] | 5.42 | 14.996 | |
| rVSV-MARV | Absent | 2 | 0 | 0 [0–0] | 0 [0–0] | 0.975 [0.558–1.39] | 3.45 | 9.752 | |
| RoNi/7.1 | rVSV-G | Induced | 2 | 7.03 × 10−5 | 0 [0–0] | 0.089 [0–0.432] | 2.47 [1.49–3.45] | 10.91 | 24.705 |
| rVSV-EBOV | Induced | 2.01 | 2.87 × 10−5 | 0 [0–0] | 0.0363 [0–0.343] | 0.685 [0.451–0.919] | 3.04 | 6.849 | |
| rVSV-MARV | Induced | 2 | 1.40 × 10−5 | 0 [0–0] | 0.0177 [0–0.257] | 1.23 [0.917–1.55] | 5.48 | 12.324 | |
| PaKiT01 | rVSV-G | Constitutive | 29.9 | .00209 | 0.00602 [0–0.019] | 8.26 × 10−8 [0–4.75 × 10−7] | 3.45 [1.07–5.84] | 6.20 | 34.516 |
| rVSV-EBOV | Constitutive | 27.9 | .00499 | 0.0478 [0–0.0958] | 4.46 × 10−8 [0–4.37 × 10−7] | 34.5 [28.7–40.2] | 18.82 | 344.821 | |
| rVSV-MARV | Induced | 2 | .00687 | 0 [0–0] | 13.1 [0–37.9] | 3.25 [0–41.3] | 8.83 | 32.452 |
-
Improvement in AIC from next best model for same cell line-virus-MOI combination. All δ-AIC are reported in Supplementary file 4.
*lci = lower and uci = upper 95% confidence interval. No confidence interval is shown for spatial β which was fixed at 10 times the estimated mean for the mean field model fits when paired with equivalent values of and ρ.
-
All other parameters were fixed at: b = 0.025 (mean field), 0.15 (spatial); α = 1/6; c = 0; μ = 1/121 (Vero), 1/191 (RoNi/7.1), and 1/84 (PaKiT01).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Vero) | Kidney (normal, epithelial, adult) | ATCC | CCL-81 | |
| Cell line (Rousettus aegyptiacus) | Kidney (normal, epithelial, adult) | (Biesold et al., 2011; Kühl et al., 2011) | RoNi/7.1 | |
| Cell line (Pteropus alecto) | Kidney (normal, epithelial, adult) | (Crameri et al., 2009) | PaKiT01 | |
| Virus strain | Replication competent, recombinant vesicular stomatitis Indiana virus expressing eGFP | (Miller et al., 2012; Wong et al., 2010) | rVSV-G | |
| Virus strain | Replication competent, recombinant vesicular stomatitis Indiana virus expressing eGFP and EBOV GP in place of VSV G | (Miller et al., 2012; Wong et al., 2010) | rVSV-EBOV | |
| Virus strain | Replication competent, recombinant vesicular stomatitis Indiana virus expressing eGFP and MARV GP in place of VSV G | (Miller et al., 2012; Wong et al., 2010) | rVSV-MARV | |
| Reagent | Hoechst 33342 Fluorescent Stain | ThermoFisher | cat #: 62249 | |
| Reagent | L-Glutamine Solution | ThermoFisher | cat #: 25030081 | |
| Reagent | Gibco HEPES | ThermoFisher | cat #: 15630080 | |
| Reagent | iTaq Universal SYBR Green Supermix | BioRad | cat #: 1725120 | |
| Commercial assay or kit | Quick RNA Mini Prep Kit | Zymo | cat #: R1054 | |
| Commercial assay or kit | Invitrogen Superscript III cDNA Synthesis Kit | ThermoFisher | cat #: 18080051 | |
| Software | MatCont (version 2.2) | (Dhooge et al., 2008) | MatCont | |
| R | R version 3.6.0 | (R Development Core Team, 2019) | R |
-
*Note that primers for R. aegyptiacus and P. alecto β-Actin, IFN-α, and IFN-β genes are listed in the Supplementary file 6.
Additional files
-
Supplementary file 1
(A) Raw proportion infectious from cell culture images.
Dataset gives raw proportion of infectious cells and time elapsed for all trials of all cell line/virus/MOI combinations, derived from image processing of binary images. (B) Raw proportion uninfectious from cell culture images. Dataset gives raw proportion of uninfectious cells and time elapsed for all trials of all cell line/virus/MOI combinations, derived from image processing of binary Hoechst-stained images. (C) Statistical mean of infectious time series for all trials of each cell line/virus/MOI experiment, from GAM fitted model incorporating random effects by trial. Data were smoothed to yield the proportion infectious per hourly timestep for each trial, and mean field mechanistic models were fit to the smoothed mean of all compiled trials for each cell line/virus/MOI combination. (D) Statistical mean of uninfectious time series for all eighteen cell line/virus/MOI experiments, from generalized linear model fit to Hoechst stain data reported on tab B. Note that these means were not used in epidemic model fitting but natural mortality rates for each cell line were derived from fitting an infection-absent model to the trajectory of susceptible decline for control trials for each cell line, as shown in Figure 1—figure supplement 7. All original raw image files, processed binary images, and image processing code are available freely for download at the following FigShare repository: DOI: 10.6084/m9.figshare.8312807.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp1-v3.xlsx
-
Supplementary file 2
Derivation of R0.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp2-v3.docx
-
Supplementary file 3
Special points from bifurcation analysis.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp3-v3.docx
-
Supplementary file 4
Optimized parameters from all deterministic model outputs and spatial approximations.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp4-v3.docx
-
Supplementary file 5
Justification for parameter increase from mean field to spatial model.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp5-v3.docx
-
Supplementary file 6
Primers for qPCR.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp6-v3.docx
-
Supplementary file 7
Detailed methods for image and image data processing.
- https://cdn.elifesciences.org/articles/48401/elife-48401-supp7-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/48401/elife-48401-transrepform-v3.pdf





