Remote control of microtubule plus-end dynamics and function from the minus-end
Figures
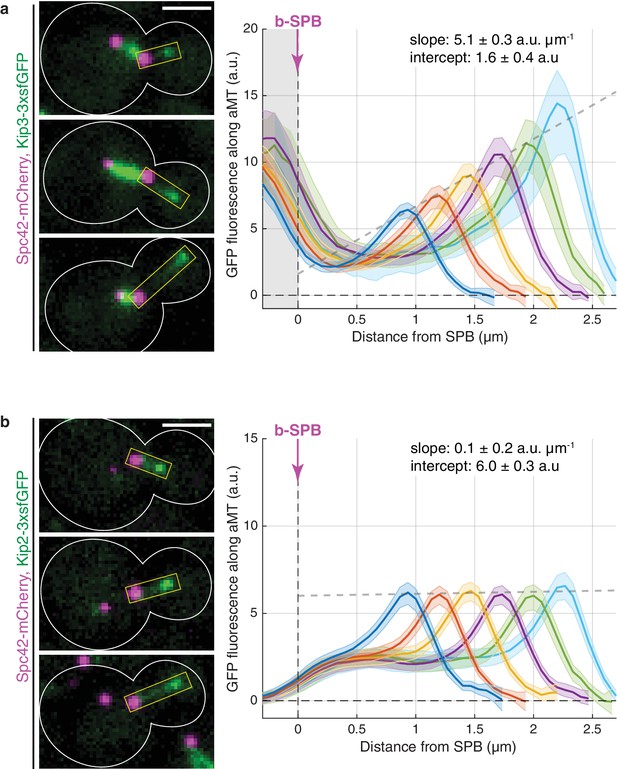
Kinesins Kip3 and Kip2 exhibit distinct localization patterns along microtubules in vivo.
(a, b) Representative images (left) and quantifications (right) of fluorescence intensities (a.u.) from endogenous Kip3-3xsfGFP (a) and Kip2-3xsfGFP (b) along preanaphase astral microtubules (aMTs; boxed areas). Signals were aligned to b-SPBs using the peak of Spc42-mCherry (magenta) intensity and binned by microtubule length (2-pixel = 266.7 nm bin size). Colored lines show mean Kip2/3-3xsfGFP fluorescence per bin and shaded areas represent 95% confidence intervals for the mean. Gray dashed lines denote weighted linear regressions for the mean GFP fluorescence on plus-ends over all bins. The area on the left of the vertical dashed line passing through x = 0 in (a) marks Kip3-3xsfGFP fluorescence inside nuclei. Scale bars, 2 µm. 10 ≤ n ≤ 130 per bin. See also Figure 1—figure supplement 1.

Functional analysis of endogenously tagged proteins and reproducibility of the kinesin distribution analysis in vivo.
(a) Defective Kip3 is synthetic lethal with dyn1∆, whereas defective Kip2 or Bik1 is synthetic lethal with bim1∆. Here we show that the expression of the Kip3-3xsfGFP, Kip2-3xsfGFP, and Bik1-3xGFP proteins does not cause synthetic growth defect or lethality with dyn1∆ or bim1∆. Spot growth assay of background cells (Spc72-GFP) and cells of indicated genotype for two days on solid YPD agar at 30°C. Spots are from exponentially growing cell samples that were sequentially diluted five-fold. (b, c) Scatterplots with fitted linear regression lines (black) between two-dimensional (2D) b-microtubule length and normalized Kip3-3xsfGFP (b) or Kip2-3xsfGFP (c) intensity on b-microtubule plus-ends (%). Kip3-3xsfGFP shows a significant positive correlation (p<0.0001, n = 183 cells) in contrast to Kip2-3xsfGFP, which shows no significant correlation (p=0.3, n = 200 cells). In addition, Kip3-3xsfGFP is nearly absent on very short microtubules, whereas Kip2-3xsfGFP is equally abundant on microtubules of all lengths. (d) Technical replicates for the quantifications of fluorescence intensities (a.u.) from endogenous Kip2-3xsfGFP along preanaphase b-microtubules. The signals were processed and presented as in Figure 1, and fluorescence intensities were normalized (see Materials and methods) to the data shown in Figure 1. Left panel: 15 ≤ n ≤ 69 per bin, right panel: 47 ≤ n ≤ 98 per bin.
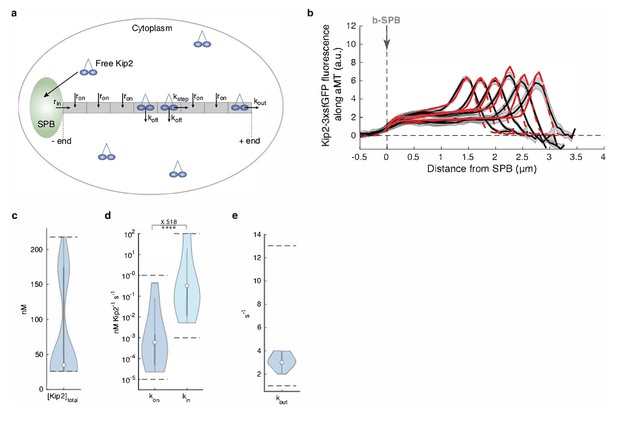
Mathematical model predicts recruitment of Kip2 to the microtubule minus-end.
(a) Schematic of the mathematical model. Free Kip2 (with concentration ) binds to the microtubule minus-end anchored at the SPB with rate if the minus-end site is free, and to any free lattice site with rate . A bound motor can detach with rate , and it can advance with rate if the next site towards the plus-end is free. At the plus end, the motor detaches with a different rate, . (b) Experimental Kip2-3xsfGFP fluorescence (a.u.) mean profile (black) and standard error (gray) with respective mean in silico model fits (red) for microtubules binned by length. Red dashed lines past plus-end and SPB indicate model extrapolations without support by data. (c) Likelihood of total Kip2 concentration estimated from fit in (b). (d) Likelihood of on rate constant and in rate constant estimated from in silico model fit in (b). Statistical significance for , ****, (p<5∙10−5) determined by sampling from the likelihood (see Appendix 1), difference in median as indicated. (e) Likelihood of out rate constant estimated from in silico model fit in (b). For in silico model parameter estimate plots (cde), median parameter values are indicated as circles, inter-quartile range () by thick gray bars and by thin gray bars. Kernel density estimates are computed from 20’000 samples from the likelihood function. Sampled parameter ranges are indicated by dashed black lines. See also Figure 2—figure supplement 2, Figure 2—figure supplement 1, and Figure 2—figure supplement 3.

Median parameter value model simulations for static and growing microtubules.
(a) In silico model simulated as in the main text, according to the measurement model section in the Supplementary Material. Briefly, the microtubule length is static and discrete microtubule lengths with 88 nm steps are considered. (b) Growing in silico microtubule model with prescribed growth rate of 0.05 s−1, corresponding to a growth speed of 0.4 nm s−1. (c) Growing in silico microtubule model with prescribed growth rate of 2.875 s−1, corresponding to the mean in vivo growth speed of 23 nm s−1 given in Figure 2—figure supplement 2d.
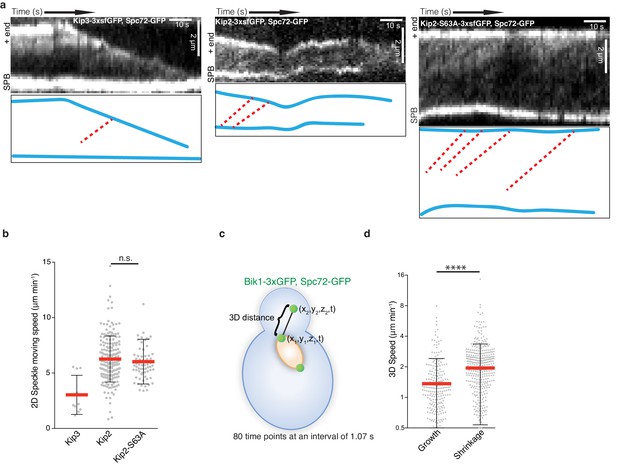
Measurements of kinesin movement speeds, as well as the speeds of b-microtubule growth and shrinkage in living cells.
(a) Representative kymographs and highlighted trajectories (red dashed lines) of Kip3-3xsfGFP (left), Kip2-3xsfGFP (middle), and Kip2-S63A-3xsfGFP (right) speckles moving along a preanaphase b-microtubule. Unlike Kip2, Kip3-3xsfGFP rarely forms obvious speckles. (b) A collection of kymographs was generated to quantify the speed of speckles moving towards microtubule plus-ends for each mutant. Kip3-3xsfGFP speckles move at a speed of 3.0 ± 1.8 µm min−1 (n = 10 speckles), Kip2-3xsfGFP at 6.3 ± 2.1 µm min−1 (n = 192), and Kip2-S63A-3xsfGFP at 6.0 ± 2.0 µm min−1 (n = 66). (c, d) The speeds of b-microtubule growth (1.4 ± 1.1 µm min−1, n = 250 phases) and shrinkage (2.0 ± 1.4 µm min−1, n = 409 phases) were derived from the 3D b-microtubule length over 85.6 s. Growth and shrinkage phases were annotated manually. All cells were cultivated and imaged at 25 °C. All values are reported as mean ± s.d.. Statistical significances were calculated using two-tailed Student’s t-test. For all panels, ****p<0.0001, n.s., not significant.
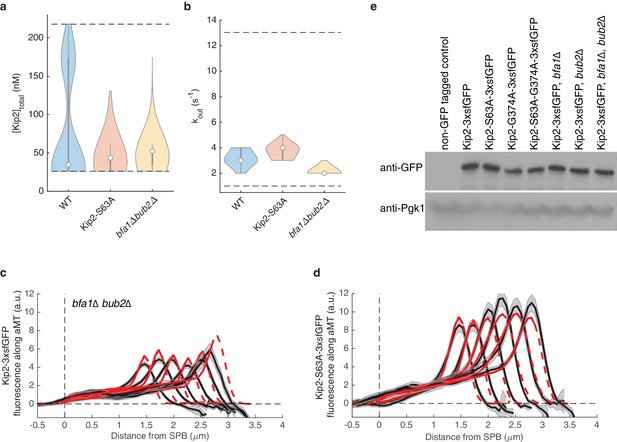
Concentration and out rate estimates for bfa1Δbub2Δ and Kip2-S63A strains.
(a) Likelihood of total Kip2 concentrations estimated from model fits in (cd) and Figure 2b. (b) Likelihood of out rate constant estimated from model fits in (cd) and Figure 2b. (c) Experimental Kip2-3xsfGFP fluorescence (a.u.) mean profile (black) and standard error (gray) in bfa1Δbub2Δ cells with respective mean of the best model fits (red). Red dashed lines past plus-end and SPB indicate model extrapolations without support by data. (d) Experimental Kip2-S63A-3xsfGFP fluorescence (a.u.) mean profile (black) and standard error (gray) with respective mean of the best model fits (red). Red dashed lines past plus-end and SPB indicate model extrapolations without support by data. (e) Western blot analysis of endogenously expressed GFP fusion proteins. Pgk1 was blotted as loading control. Lysates were prepared from cycling cells of indicated genotype.
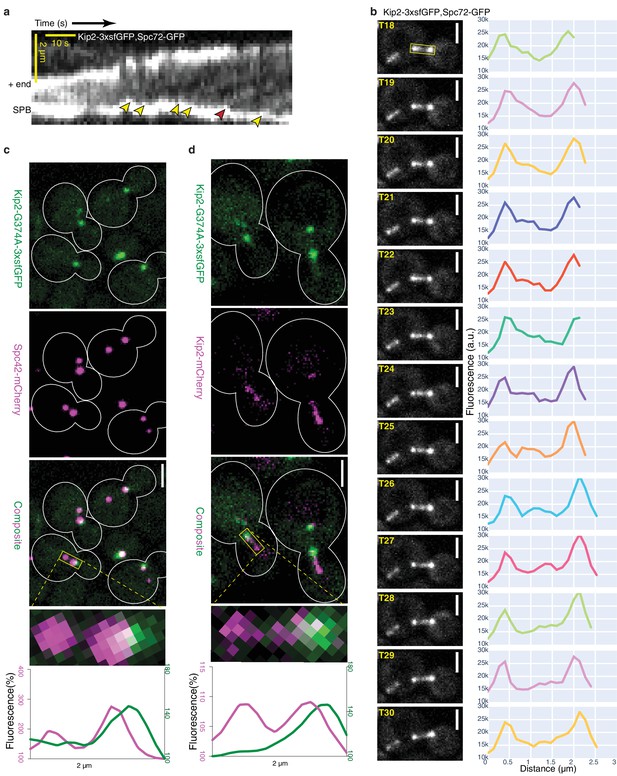
Kip2 initiates its runs from SPBs or their vicinity.
(a) Representative experimental kymograph showing Kip2-3xsfGFP speckles departing (yellow arrows) from the SPB, visualized with Spc72-GFP. The red arrow marks a Kip2-3xsfGFP speckle that moves towards the part of the microtubule which moves out of focus. (b) Images of Video 1 from time frame 18 to 30 and line scan analysis of the b-microtubule (boxed area) showing a Kip2-3xsfGFP speckle that departs from the SPB (T18–T19), moves along the microtubule shaft (T20–T29), and arrives at the plus-end (T30). (c) Representative images of preanaphase cells expressing the endogenous ATPase deficient protein Kip2-G374A-3xsfGFP (green) and Spc42-mCherry (magenta). Close-up of a mitotic spindle (boxed area, 2 µm long) and line scan analysis of this area are shown on the right. Scale bars, 2 µm. (d) Representative images of preanaphase heterozygous diploid cells expressing Kip2-G374A-3xsfGFP (green) and the wild type protein Kip2-mCherry (magenta). Close-up (boxed area, 2 µm long) and line scan analysis as in (b). Scale bars, 2 µm. For the line scan analysis in (b,c), GFP (green) and mCherry (magenta) fluorescence intensities were normalized to their background levels, respectively. See also Video 1, Figure 2—figure supplement 2a, Figure 3—figure supplement 1 and Figure 3—figure supplement 2.

Sequence alignment of various kinesin motor domains focused on the switch-2 motif (DxxGxE, highlighted in bold) (Marx et al., 2009) that is essential for the ATP hydrolysis.
The yeast (sc) kinesins Kip2 (UniProt ID: P28743), Kip1 (UniProt ID: P28742) and Kip3 (UniProt ID: P53086) were aligned with the human (hs) kinesins CENPE (UniProt ID: Q02224), Kif5a (UniProt ID: Q12840) and Kif2C (UniProt ID: Q99661) to illustrate conservation across species and kinesin families. The crucial glycine residue, which was mutated to alanine for Kip2, is highlighted in blue.
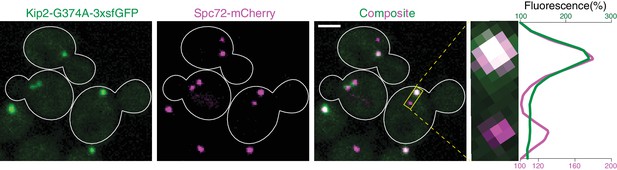
Kip2-G374A-3xsfGFP colocalizes with Spc72-mCherry.
Representative images of cells coexpressing Kip2-G374A-3xsfGFP (green) with the SPB outer plaque component Spc72-mCherry (magenta). A close-up of the boxed area (preanaphase spindle) is shown on the right, accompanied with the line scan analysis along the boxed area. Scale bar, 2 µm.
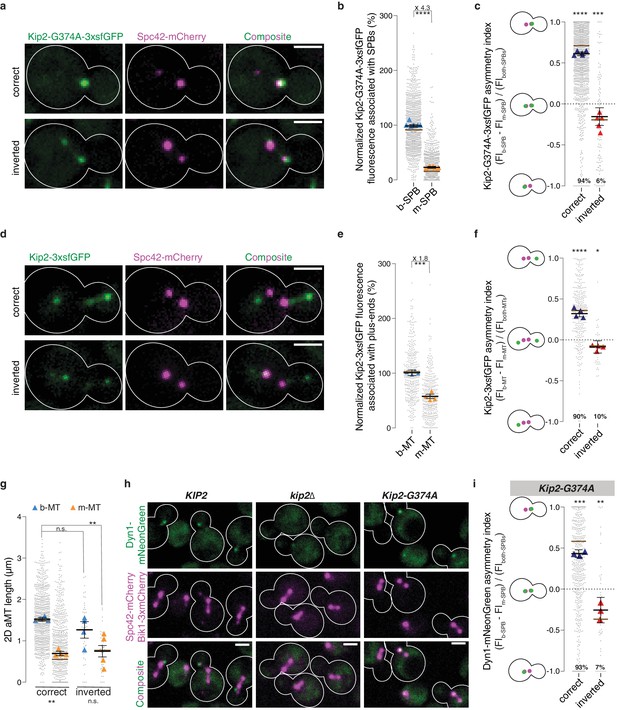
Biased microtubule growth and dynein distribution correlate with SPB dependent Kip2 recruitment.
(a) Localization of Kip2-G374A-3xsfGFP (green) in preanaphase cells with correctly oriented (top) and inverted (bottom) SPBs. The orientation of the SPBs is visualized with Spc42-mCherry (magenta). (b) Relative Kip2-G374A-3xsfGFP fluorescence (%) associated with b- and m-SPBs in cells shown in (a) (999 cells from n > 3 independent clones or technical replicates). (c) Quantification of asymmetry index for Kip2-G374A-3xsfGFP distribution (fluorescence intensity: (FIb-SPB – FIm-SPB) / FIboth-SPBs) between SPBs in cells in (a) with correctly orientated (1407 cells) and inverted SPBs (97 cells). Statistical significances of difference from zero were tested with one-way ANOVA. (d) Localization of Kip2-3xsfGFP (green) in preanaphase cells carrying both b- and m- microtubules, with correctly oriented (top) and inverted (bottom) SPBs. (e) Normalized fluorescence (%) of Kip2-3xsfGFP associated with plus-ends of b- and m-microtubules in preanaphase cells in (d) (347 cells from n > 3 independent clones or technical replicates). (f) Quantification of asymmetry index for Kip2-3xsfGFP accumulation at m- and b-microtubule plus-ends (fluorescence intensity: (FIb-MT – FIm-MT) / FIboth-MTs) in preanaphase cells in (d) with correctly orientated (314 cells) and inverted SPBs (33 cells). Statistical significances of difference from zero were tested with one-way ANOVA. (g) Measurements of two-dimensional (2D) b- and m- microtubule lengths (μm) in cells with correctly orientated (758 cells) and inverted SPBs (40 cells) using Kip2-3xsfGFP and Spc42-mCherry as microtubule plus- and minus-end markers, respectively. In case of no visible astral microtubule, the microtubule length was set to 0 μm. (h) Localization of Dyn1-mNeonGreen (green) in cells of indicated genotype. Spindles and aMTs are visualized with Spc42-mCherry and Bik1-3xmCherry (magenta). (i) Quantification of asymmetry index for Dyn1-mNeonGreen distribution (fluorescence intensity: (FIb-SPB – FIm-SPB) / FIboth-SPBs) in Kip2-G374A cells with correctly orientated (641 cells) and inverted SPBs (47 cells). Note that in 13.4% (106 out of 794) of cells, no detectable Dyn1-mNeonGreen accumulated on preanaphase spindles; these cells were not included in the analysis. Statistical significances of difference from zero were tested with one-way ANOVA. For all panels, means with 95% confidence intervals are shown in black; median values shown as brown bar; data were acquired from at least three independent clones and technical replicates. Relative GFP fluorescence was obtained by normalizing to the mean GFP fluorescence associated with b-SPBs or plus-ends of b-microtubules, as indicated. The average value of each clone or technical replicate is plotted as triangle. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s., not significant. Statistical significances were calculated using one-way ANOVA (b,c,e,f,g,i). Source data for these panels are available in Supplementary file 1. Scale bars, 2 µm.
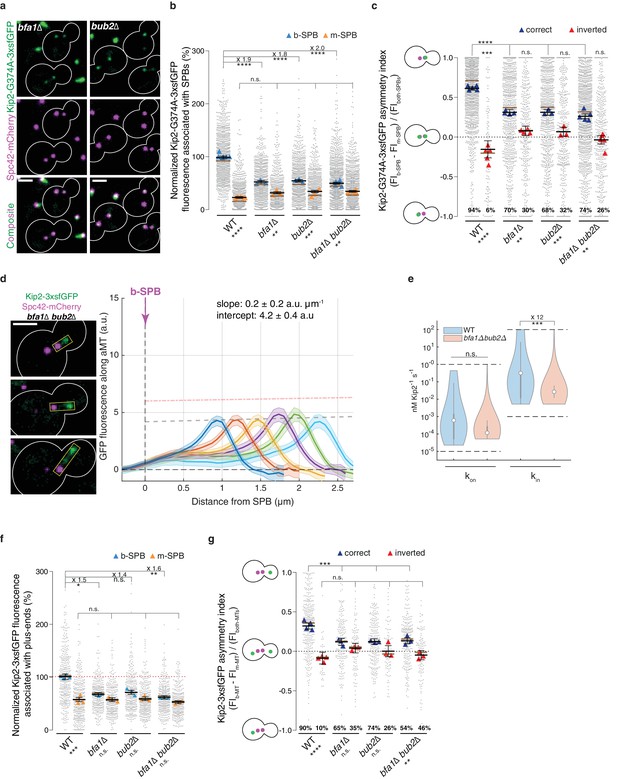
Bfa1 and Bub2 promote Kip2 run initiation from bud-directed SPBs.
(a) Representative images of Kip2-G374A-3xsfGFP (green) in preanaphase cells of indicated genotype. Spindles are visualized with Spc42-mCherry (magenta). See Figure 3c for control. (b) Normalized Kip2-G374A-3xsfGFP fluorescence (%) associated with b- (blue) and m-SPBs (yellow) in cells shown in (a) (n > 3 independent clones or technical replicates,>701 cells per genotype). (c) Quantification of asymmetry index for Kip2-G374A-3xsfGFP distribution (fluorescence intensity: (FIb-SPB – FIm-SPB) / FIboth-SPBs) between SPBs in cells in (b) with correctly orientated (blue) and inverted (red) SPBs. For cells with inverted SPBs, statistical significances of difference from zero were tested with one-way ANOVA. (d) Representative images (left) and quantifications (right) of fluorescence intensities (a.u.) from endogenous Kip2-3xsfGFP along preanaphase aMTs (boxed areas) in bfa1∆bub2∆ cells. The graph format is the same as in Figure 1. 45 ≤ n ≤ 99 per bin. The pink dashed line denotes the weighted linear regressions for the mean GFP fluorescence on plus-ends in wild-type cells. See Figure 5—figure supplement 1 for more details. (e) In silico likelihood of on rate constant and in rate constant estimated from model fits to Kip2-3xsfGFP distribution in wt and bfa1∆bub2∆ cells. Statistical significance for (***, p=8∙10−4, was determined by sampling from the likelihood for (see Appendix 1), difference in median as indicated. Graph as in Figure 2d. (f) Normalized Kip2-3xsfGFP fluorescence (%) associated with microtubule plus-ends in preanaphase cells carrying both b- (blue) and m- (yellow) microtubules (n > 3 independent clones or technical replicates,>275 cells per genotype). See Figure 5—figure supplement 1a for representative images. (g) Quantification of asymmetry index for Kip2-3xsfGFP accumulation at m- and b-microtubule plus-ends (fluorescence intensity: (FIb-MT – FIm-MT) / FIboth-MTs) in preanaphase cells for data in (f) with correctly orientated (blue) and inverted (red) SPBs. Graphs and statistical analysis are as in Figure 4 (b,c,e,f,g and i) unless otherwise indicated. Statistical significances within each genotype are marked at the bottom. Scale bars, 2 µm. See also Figure 5—figure supplement 1.
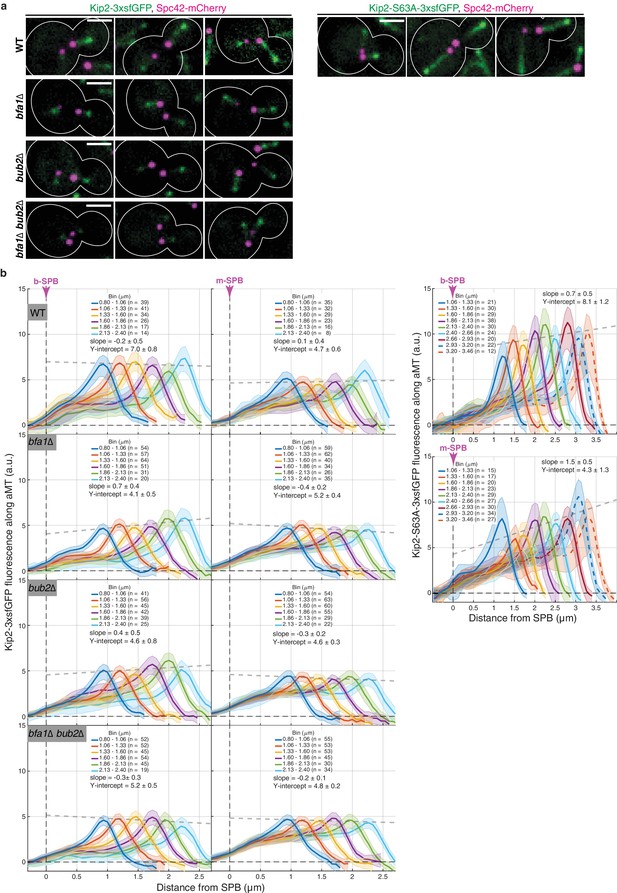
Kip2 localization patterns along microtubules in cells with both b- and m- microtubules.
(a) Representative images of Kip2-3xsfGFP (left) and Kip2-S63A-3xsfGFP (right) distribution along preanaphase aMTs in cells of indicated genotype. Scale bars, 2 µm. (b) Distribution of Kip2-3xsfGFP (left) and Kip2-S63A-3xsfGFP (right) fluorescence (a.u.) in cells of indicated genotype along b- and m- microtubules as a function of microtubule length (µm). Colored lines show mean GFP fluorescences per length bin and shaded areas 95% confidence intervals for the mean. Gray dashed lines denote weighted linear regression for the mean GFP fluorescence on microtubule plus-ends over all bins, with slopes, intercepts, and standard errors as detailed. The numbers of analyzed preanaphase cells are marked for each bin.
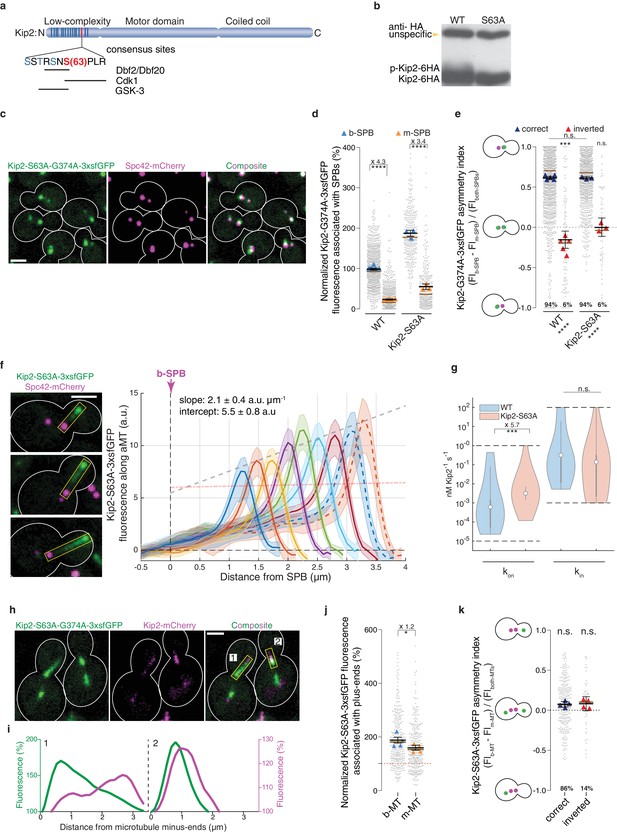
Phosphorylation of its N-terminus prevents Kip2 from landing along microtubules.
(a) Scheme of Kip2 protein and its disordered N-terminus with residue serine 63 (red) and kinase consensus sites (blue). (b) Western blot analysis of endogenously expressed Kip2-6HA and Kip2-S63A-6HA. Lysates were prepared from cycling cells of indicated genotype. (c) Representative images of Kip2-S63A-G374A-3xsfGFP (green) in preanaphase cells. Spindles are visualized with Spc42-mCherry (magenta). See Figure 3b for control. (d) Normalized Kip2-S63A-G374A-3xsfGFP fluorescence (%) associated with b- (blue) and m-SPBs (yellow) in cells shown in (c) (n > 3 independent clones or technical replicates,>314 cells per genotype). (e) Quantification of asymmetry index for Kip2-S63A-G374A-3xsfGFP distribution (fluorescence intensity: (FIb-SPB – FIm-SPB) / FIboth-SPBs) between SPBs for data in (d) with correctly orientated (blue) and inverted (red) SPBs. (f) Representative images (left) and quantifications (right) of fluorescence intensities (a.u.) from endogenous Kip2-S63A-3xsfGFP along preanaphase aMTs (boxed areas). The graph format is the same as those in Figure 1. 29 ≤ n ≤ 55 per bin. The pink dashed line denotes the weighted linear regression for the mean GFP fluorescence on plus-ends in wild-type cells. See Figure 5—figure supplement 1 for more details. (g) In silico likelihood of on rate constant and in rate constant estimated from model fits to Kip2-3xsfGFP data in wt and Kip2-S63A-3xsfGFP cells. Statistical significance for (***, p<2∙10−4) was determined by sampling from the likelihood for (see Appendix 1), difference in median as indicated. Graph as in Figure 2d. (h) Representative images of preanaphase heterozygous diploid cells expressing the endogenous ATPase deficient protein Kip2-S63A-G374A-3xsfGFP (green) and the wild type protein Kip2-mCherry (magenta). (i) Line scan analysis along the numbered microtubules shown in (h). As in Figure 3b, GFP (green) and mCherry (magenta) fluorescence intensities were normalized to their background levels, respectively. (j) Normalized Kip2-S63A-3xsfGFP fluorescence (%) associated with microtubule plus-ends in preanaphase cells carrying both b- (blue) and m- microtubules (yellow) (305 cells from n > 3 independent clones). Values were normalized to Kip2-3xsfGFP fluorescence associated with b-microtubules in wild-type cells, denoted as red dotted line in the graph. See Figure 5—figure supplement 1a for representative images. (k) Quantification of asymmetry index for Kip2-S63A-3xsfGFP accumulation on microtubule plus-ends (fluorescence intensity: (FIb-MT – FIm-MT) / FIboth-MTs) for data in (j) with correctly orientated (blue) and inverted (red) SPBs. Graphs and statistical analysis are like those in Figure 4 (b,c,e,f,g and i). Statistical significances within each genotype are marked at the bottom of graphs. Source data for these panels are available in Supplementary file 1. Scale bars, 2 µm. See also Video 3, Figure 5—figure supplement 1 and Figure 6—figure supplement 1.
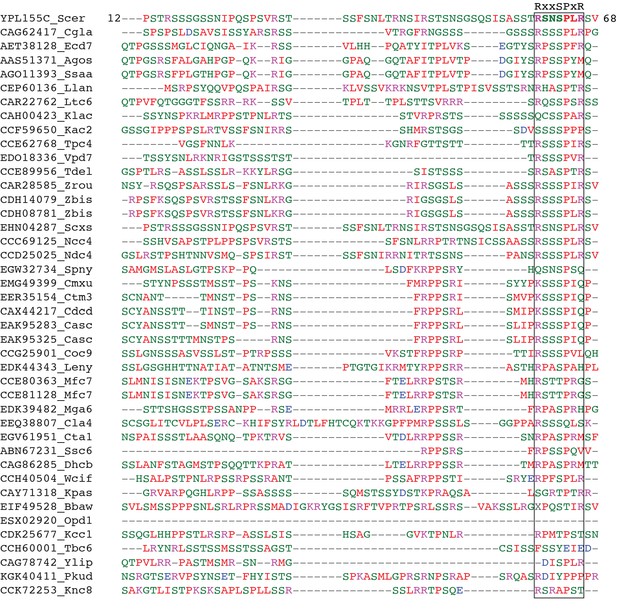
Conservation of Kip2’s low complexity N-terminus.
Protein sequence alignment of the Kip2’s low complexity N-terminus and its homologs among Saccharomycetales. The tandem kinase consensus site (RxxSPxR, in which x represents any amino acid residue) covering Serine 63, as well as the richness in serine and threonine residues are highly conserved. Alignment was generated using MUSCLE (Edgar, 2004) and manually curated.
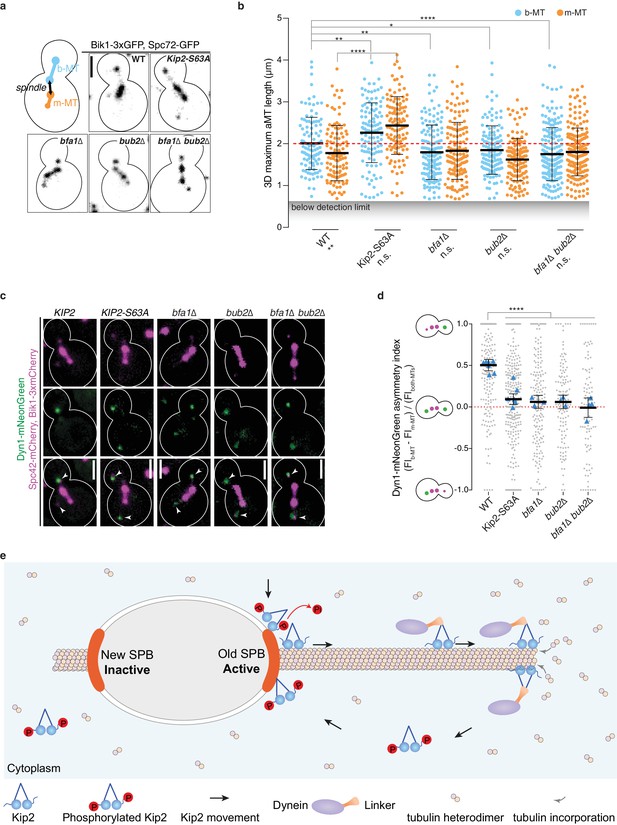
SPB dependent recruitment of Kip2 specifies aMT size and dynein distribution.
(a) Illustration and representative images of preanaphase cells of indicated genotype carrying both b- and m-microtubules using Bik1-3xGFP and Spc72-GFP as microtubule plus- and minus-end markers, respectively. Scale bars, 2 µm. (b) Quantification of the three-dimensional (3D) average maximum length (µm) of b- (blue) and m-microtubules (yellow) over the recording time (85.6 s, see Materials and methods) from the cells shown in (a). Means with 95% confidence intervals are shown in black, n > 127 cells per genotype. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s., not significant. Statistical significances were calculated using two-tailed Student’s t-test. Source data are available in Supplementary file 1. (c) Localization of Dyn1-mNeonGreen (green) in preanaphase cells of indicated genotype carrying both aMTs. Spindles and aMTs are visualized with Spc42-mCherry and Bik1-3xmCherry (magenta). Scale bars, 2 µm. (d) Quantification of asymmetry index (fluorescence intensity: (FIb-MT – FIm-MT) / FIboth-MTs) of Dyn1-mNeonGreen accumulation on microtubule plus-ends in cells shown in (c) (>94 cells per genotype). Means with 95% confidence intervals are shown in black; data were acquired from at least three independent clones or technical replicates. The average value of each clone or technical replicate is plotted as triangle. ****p<0.0001, Statistical significances were calculated using one-way ANOVA. Source data for this panel are available in Supplementary file 1. (e) Illustration of the proposed remote-control mechanism (see main text for details). See also Figure 7—figure supplement 1 and Figure 7—figure supplement 2.
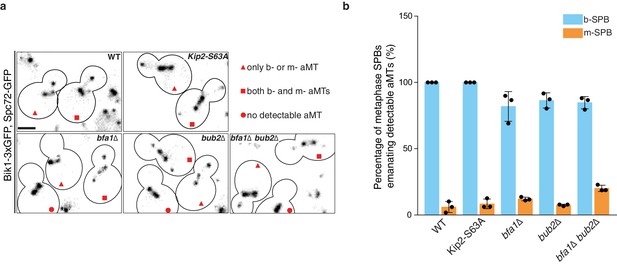
Astral microtubule organization in preanaphase yeast cells.
(a) Representative images of preanaphase cells of indicated genotype expressing Bik1-3xGFP and Spc72-GFP as aMT plus- and minus-end markers, respectively. Cells that make only one aMT are denoted with triangles, those carrying both b- and m- aMTs are marked with squares, and cells failing to build visible aMTs are marked with circles. Scale bars, 2 µm. (b) Quantification of detectable aMTs emanating from b-SPB (blue) and m-SPB (yellow) in preanaphase cells of indicated genotype (n = 3 independent clones,>300 cells per genotype). Due to the resolution limit of the microscope, an astral microtubule (aMT) was defined as detectable when its length was above five pixels (666.7 µm).

Pairwise Dyn1-mNeonGreen distribution between b- and m-microtubules in control and Kip2-S63A cells.
Pairwise relative Dyn1-mNeonGreen fluorescence (%) associated with b- and m-microtubule plus-ends in cells shown in Figure 7c (n > 94 cells per genotype). Each blue line connects two dots that were measurements of Dyn1-mNeonGreen fluorescence from one cell. Means with standard deviations are shown in red.
Videos
A representative time-lapse movie shows Kip2-3xsfGFP molecules appearing as speckles from the SPB and moving along the cytoplasmic microtubule towards its plus-end.
All Kip2-3xsfGFP speckles reach the plus-end. The movie consists of 80 frames that were taken every 1.07 s and the frame speed is sped up by 3-fold for better visualization. Scale bar, 2 µm.
A representative time-lapse movie shows that Kip3-3xsfGFP molecules are nearly absent from the minus-end of the cytoplasmic microtubule and accumulate along the shaft to reach the maximum intensity on the plus-end.
The movie consists of 80 frames that were taken every 1.07 s and the frame speed is sped up by 3-fold for better visualization. Scale bar, 2 µm.
A representative time-lapse movie shows that unlike wildtype molecules, Kip2-S63A-3xsfGFP molecules accumulate along the cytoplasmic microtubule shaft length-dependently.
Also, these speckles move towards and reach the plus-end. The movie consists of 80 frames that were taken every 1.07 s and the frame speed is sped up by 3-fold for better visualization. Scale bar, 2 µm.
Additional files
-
Supplementary file 1
Source data for Figure 4bcdefgi, Figure 5bceg, Figure 6dejk, Figure 7bd, Figure 1—figure supplement 1, Figure 2—figure supplement 2bd, and Figure 7—figure supplement 1b.
- https://doi.org/10.7554/eLife.48627.022
-
Supplementary file 2
Strains used in this study.
- https://doi.org/10.7554/eLife.48627.023
-
Supplementary file 3
Stochastic simulation propensities and change vectors.
Git code repository: https://gitlab.com/csb.ethz/Kip2-SPB-Profile-Manuscript (Chen, 2019; copy archived at https://github.com/elifesciences-publications/Kip2-SPB-Profile-Manuscript).
- https://doi.org/10.7554/eLife.48627.024