Function of hTim8a in complex IV assembly in neuronal cells provides insight into pathomechanism underlying Mohr-Tranebjærg syndrome
Figures
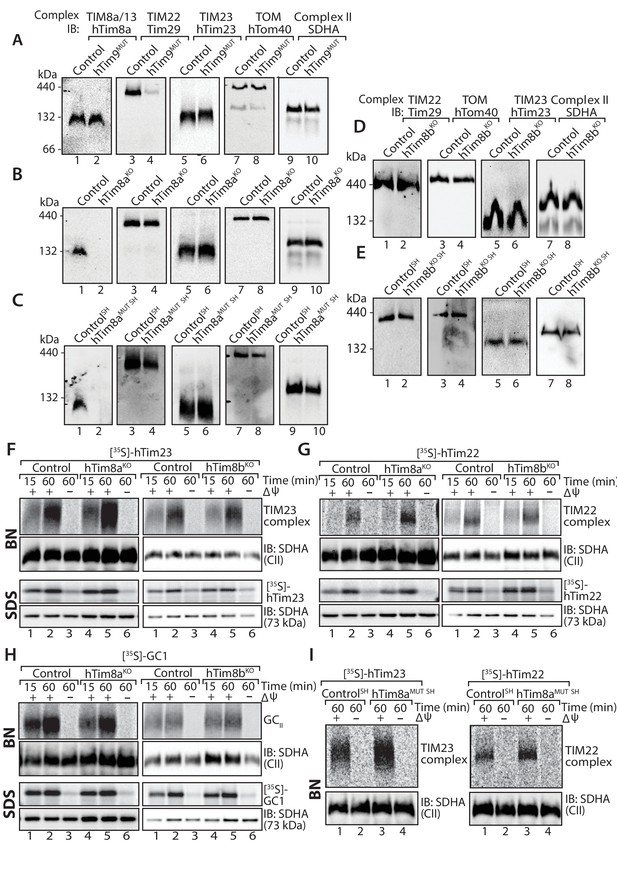
Cells lacking hTim8a have no defects in the TIM22, TIM23 or carrier biogenesis pathway.
(A–E) Mitochondria were isolated from control, (A) hTim9MUT HEK293 cells, (B) hTim8aKO HEK293 cells, (C) hTim8aMUT SH-SY5Y cells, (D) hTim8bKO HEK293 cells, or (E) hTim8bKO SH-SY5Y cells prior to solubilisation in 1% digitonin-containing buffer. Mitochondrial lysates were subjected to Blue-Native electrophoresis prior to immunoblotting using the indicated antibodies. (F–H) [35S]-hTim23, [35S]-hTim22 or [35S]-GC1 were incubated with mitochondria isolated from control and hTim8aKO or hTim8bKO) HEK293 cells for the indicated time in the absence or presence of a mitochondrial membrane potential (ΔΨ) prior to Proteinase K treatment. Samples were separated by SDS-PAGE or solubilised in 1% digitonin-containing buffer and separated by BN-PAGE and visualised using autoradiography. (I) Mitochondria isolated from control SH-SY5Y and hTim8aMUT SH cells were incubated with [35S]-hTim23 or [35S]-hTim22 for 60 min in the presence or absence of membrane potential (ΔΨ) before Proteinase K treatment. Mitochondria were reisolated and solubilised in digitonin prior to BN-PAGE and subsequent immunoblotting.
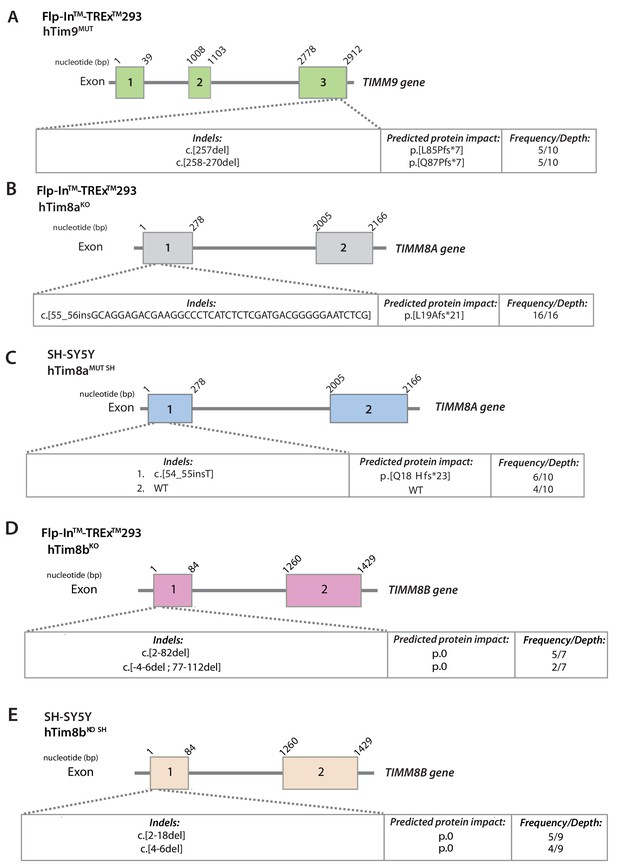
Generation of CRISPR/Cas9 genome-edited cell lines.
(A–E) Schematic representation of the CRISPR/Cas9 editing system used to generate (A) hTim9MUT (MUT = mutant) in Flp-In T-REx HEK293 cells, (B) hTim8aKO (KO = knock out) in Flp-In T-REx HEK293 cells, (C) hTim8aMUT (MUT = mutant) in SH-SY5Y cells (D) hTim8bKO in Flp-In T-REx HEK293 cells or (E) hTim8bKO (KO = knock out) in SH-SY5Y cells.
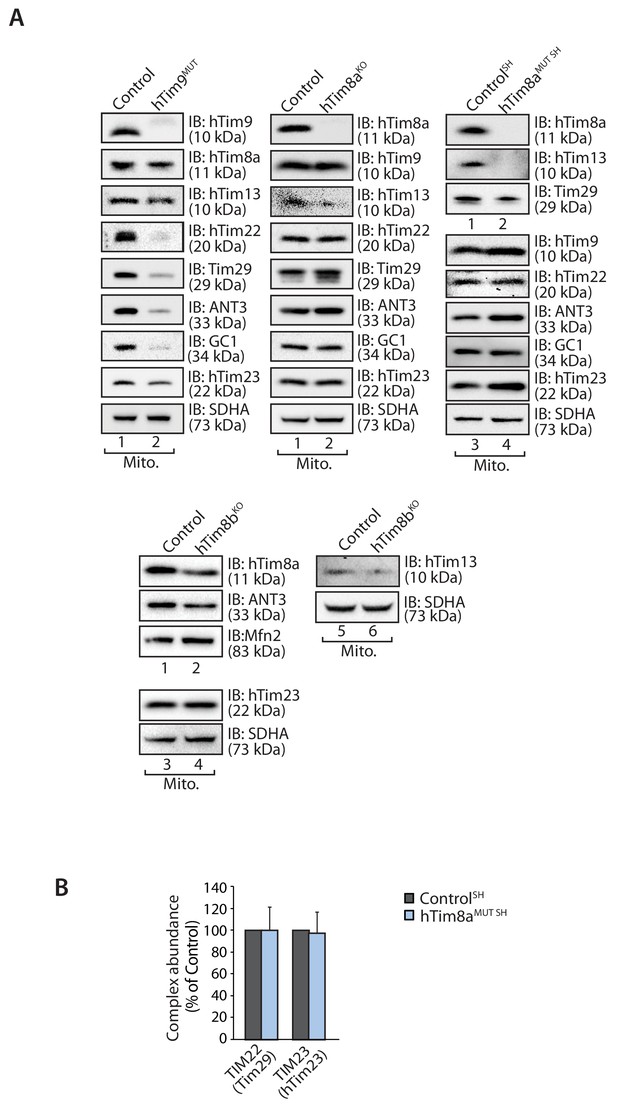
Loss of hTim8a or hTim8b has no defect on TIM or carrier biogenesis.
(A) Mitochondria isolated from control and hTim9MUT, hTim8aKO, hTim8aMUT SH or hTim8bKO SH cells, were subjected to SDS-PAGE analysis and immunoblotted using the indicated antibodies. (B) Relative abundance of TIM22 complex and TIM23 complex in control and hTim8aMUT SH-SY5Y mitochondria (refer to the BN-PAGE/western blot analysis in Figure 1C) were quantified and tabulated as mean ± SD (n = 3).
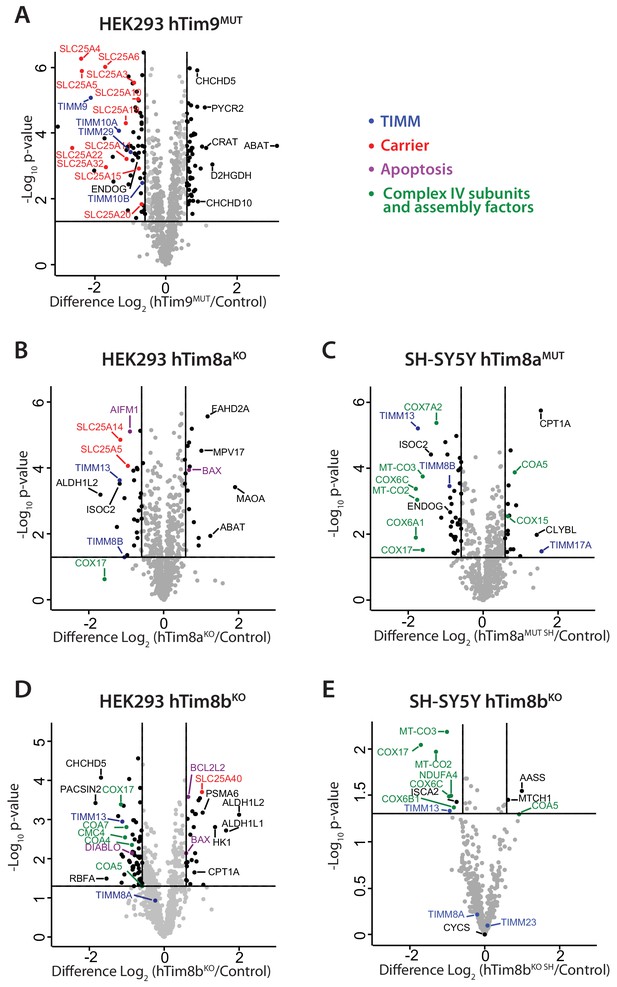
Loss of hTim8a shows cell-type specific consequences on the mitochondrial proteome.
(A–E) Mitochondria were isolated from control and (A) hTim9MUT HEK293, (B) hTim8aKO HEK293, (C) hTim8aMUT SH-SY5Y, (D) hTim8bKO HEK293, or (E) hTim8bKO SH-SY5Y cells and subjected to label-free quantitative mass spectrometry analyses. Volcano plots showing relative levels of proteins in knock-out or mutant cells compared to control cells. n = 3 biological replicates. Significantly altered proteins are located outside the line (p-value<0.05): TIMM proteins(blue), carrier proteins of the SLC25 family (red), apoptotic-related proteins (purple) and complex IV subunits and assembly factors (green) are indicated.
-
Figure 2—source data 1
Label-free quantitiative mass spectrometry on mitochondria isolated from: hTim9MUT, hTim8aKO, hTim8aMUT SH,hTim8bKO and hTim8bKO SH.
- https://cdn.elifesciences.org/articles/48828/elife-48828-fig2-data1-v3.xlsx
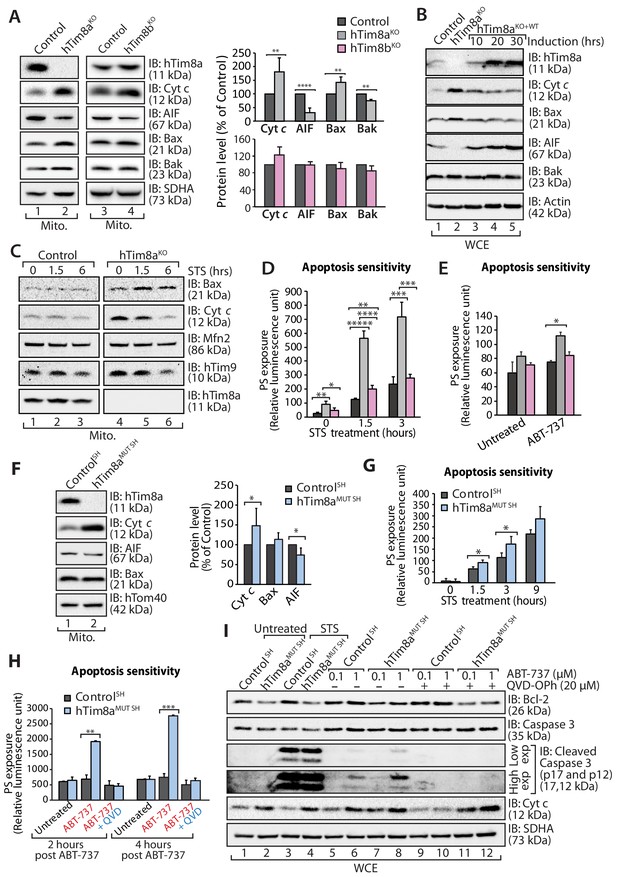
Lack of hTim8a, but not hTim8b, sensitises cells to apoptotic-cell death.
(A) Mitochondrial lysates from control and hTim8aKO (left panel) or hTim8bKO (right panel) HEK293 cells were analysed by SDS-PAGE and western blotting. Relative protein levels of cytochrome c (Cyt c), apoptosis-inducing factor (AIF), Bax and Bak were quantified and tabulated as mean ± SD (n = 3). **, p<0.01; ****, p<0.0001. (B) Cell lysates from control, hTim8aKO and hTim8aKO cells re-expressing hTim8a (hTim8aKO+WT) were analysed using SDS-PAGE and immunoblotting with the indicated antibodies. (C) Control and hTim8aKO cells were treated with staurosporine (STS; 1.5 μM) for 0, 1.5 or 6 hours prior to mitochondrial isolation and analysis by SDS-PAGE. (D and E) Control, hTim8aKO and hTim8bKO HEK293 cells were (D) treated with staurosporine (STS; 1.5 μM) for 0, 1.5 or 3 hours, or (E) incubated with ABT-737 (0.1 μM) for 0 or 2 hours. The rate of apoptosis was calculated by measuring phosphatidylserine (PS) exposure to the outer leaflet of plasma membrane (relative luminescence unit). n = 4 (STS treatment); n = 3 (ABT-737 treatment); mean ± SD; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001; *****, p<0.00001. (F) Mitochondria isolated from control SH-SY5Y and hTim8aMUT SH cells were analysed by SDS-PAGE and immunoblotting. Graph shows the relative levels of hTim23, Bax, Cyt c and AIF quantified and represented as mean ± SD (n = 5). *, p<0.05. (G) Apoptotic sensitivity of control and hTim8aMUT SH cells was measured following staurosporine (STS; 1.5 μM) treatment by assessing phosphatidylserine (PS) exposure (relative luminescence unit). n = 4, mean ± SD; *, p<0.05. (H) Control and hTim8aMUT SH cells was either: (i) left untreated, (ii) treated with ABT-737 (0.1 μM) or (iii) pretreated with QVD-OPh (20 μM) for 20 min prior to ABT-737 treatment, for 2 or 4 hr prior to measuring cellular apoptotic sensitivity by assessing phosphatidylserine (PS) exposure (relative luminescence unit). n = 3, mean ± SD; **, p<0.01; ***, p<0.001. (I) Control and hTim8aMUT cells were either (i) left untreated, (ii) treated with STS (1.5 μM) or (iii) treated with ABT-737 (0.1 or 1 μM) with or without preincubation with QVD-OPh. Cell lysates were harvested following these treatments for SDS-PAGE and immunoblot analyses using the indicated antibodies.
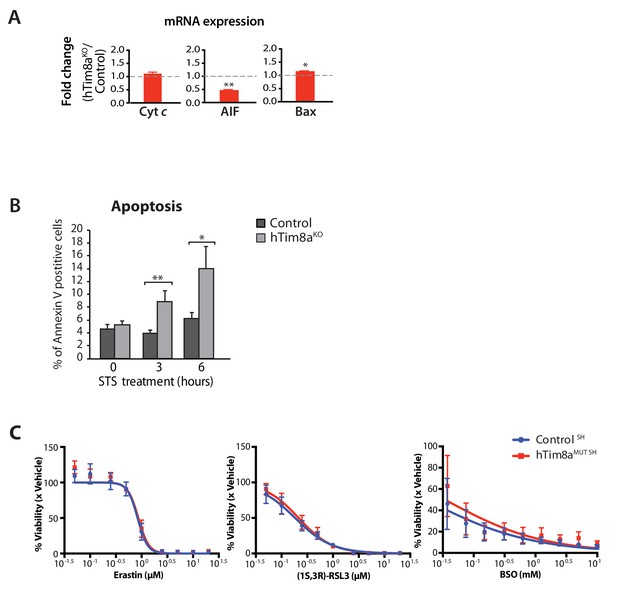
Cells lacking fucntional hTim8a are not sensitised to ferroptosis-induced cell death, but intrinsic cell death.
(A) Relative fold change of mRNA expression of Cyt c, AIF and Bax in hTim8aKO compared to control HEK293 were quantified and tabulated as mean ± SEM (n = 3). *, p<0.05; **, p<0.01. (B) Control and hTim8aKO HEK293 cells were treated with STS for the indicated time prior to harvesting and staining with Annexin V for FACS analysis. The % of Annexin V positive (apoptotic) cells were quantified from a total population of ~20,000 cells and were represented as mean ± SD, n = 3 biological replicates. *, p<0.05; **, p<0.01. (C) Control and hTim8aMUT SH-SY5Y cells were treated with ferroptosis inducers at the indicated concentrations: Erastin (SLC7A11 inhibitor), (1S,3R)-RSL3 (GPX4 inhibitor) or BSO (γ-GCS inhibitor), for 72 hr to induce ferroptosis. The cellular viability following drug treatment was quantified using alamarBlue assay and represented as mean ± SEM, n = 4 biological replicates.
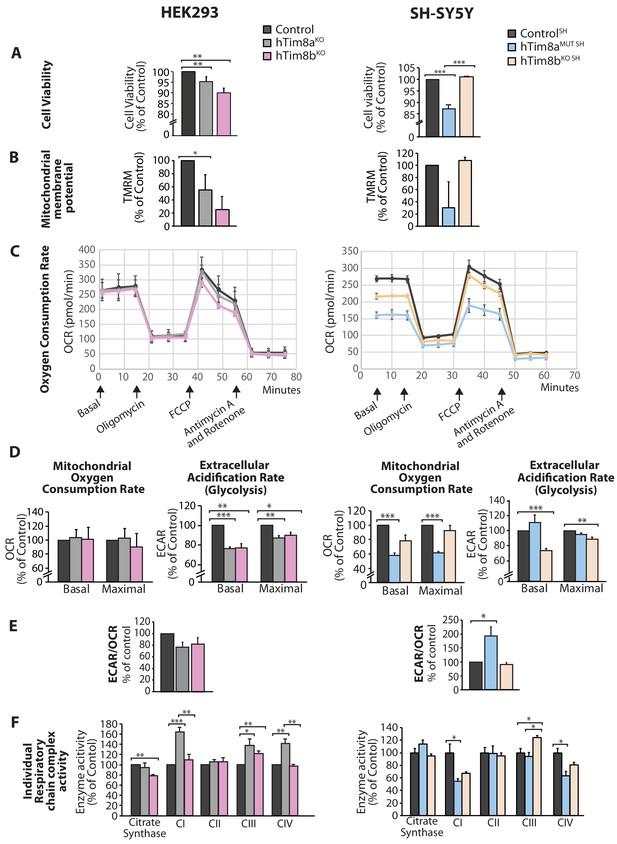
Cellular dysfunction in cells lacking hTim8a or hTim8b.
(A) Cell viability of control and hTim8aKO or hTim8bKO HEK293 cells (left panel) or hTim8aMUT SH or hTim8bKO SH cells (right panel) was measured using trypan blue staining. Cells were seeded at the same confluency and 24 hours later were stained with trypan blue for cell counting. Viable cells were calculated as a percentage (1- [dead cells [stained blue]/total number of cells] X 100 = % viable cells). Data represents mean ± SD (n = 3). **, p<0.01, ***, p<0.001. (B) Mitochondrial membrane potential in control and hTim8aKO or hTim8bKO HEK293 cells (left panel) or hTim8aMUT SH or hTim8bKO SH cells (right panel) were quantified using TMRM uptake. Data represents mean ± SD (n = 3). *, p<0.05. (C) Mitochondrial oxygen consumption rate of control and hTim8aKO/hTim8bKO HEK293 cells (left panel) or hTim8aMUT or hTim8bKO SH-SY5Y cells (right panel) were measured using a Seahorse analyser. Oligomycin, FCCP, Antimycin A and Rotenone were added at the indicated time to the cells to allow for the measurement of basal, maximal and non-mitochondrial respiration rate. Error bars represent SEM (n=3). (D) The basal and maximal mitochondrial oxygen consumption rate (OCR; left panel) or extracellular acidification rate (ECAR; right panel) of hTim8a or hTim8b-edited cells relative to control were calculated and represented as mean ± SEM (n = 3). *, p<0.05, **, p<0.01, ***, p<0.001. (E) The ratio of ECAR/OCR of control and hTim8a or hTim8b-edited cells were calculated and were subsequently normalised to control and represented as mean ± SEM (n = 3). *, p<0.05. (F) The relative activity of individual respiratory chain complexes: Complex I-IV and citrate synthase in control and hTim8aKO or hTim8bKO HEK293 cells (left panel) or hTim8aMUT or hTim8bKO SH-SY5Y cells (right panel) were measured and represented as mean ± SEM (n = 3). *, p<0.05, **, p<0.01, ***, p<0.001.
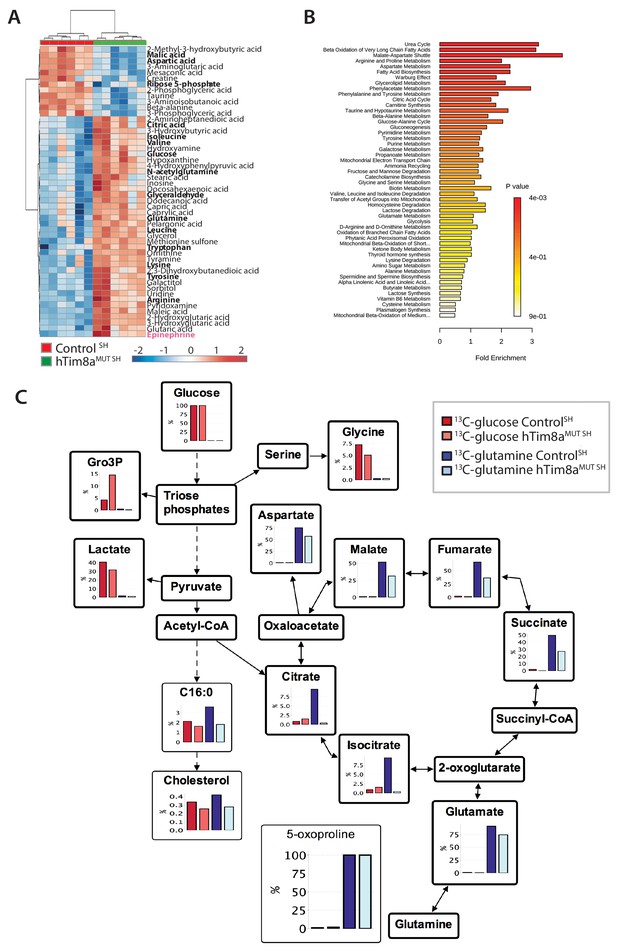
Metabolic rewiring in hTim8aMUT SH-SY5Y cells.
(A) Relative abundance of significantly affected metabolites in hTim8aMUT compared to control SH-SY5Y cells. Heatmap representing the top 50 most affected metabolites extracted from control and hTim8aMUT SH cells (p<0.05). Data represents values from six independent biological replicates of control and hTim8MUT cells and fold changes are color-coded as indicated. See also Figure 5—source data 1 and Figure 5—figure supplement 2. (B) Top 50 most affected cellular metabolic pathways in control and hTim8aMUT SH cells were identified and represented as a clustered bar chart with fold enrichment indicated. The p-value of each of the metabolic pathways are color-coded as indicated. See also Figure 5—source data 1 and Figure 5—figure supplement 2. (C) Percentage (%) of U-13C-glucose (indicated by red bars) or U-13C-glutamine (indicated by blue bars) labelled intracellular metabolites following a 2 hr incubation of control and hTim8aMUT SH cells in 13C-glucose or 13C-glutamine labelled media. Data represents the mean of 3 independent experiments and are mapped onto their respective metabolic pathway. See also Figure 5—source data 2.
-
Figure 5—source data 1
Untargeted metabolomics profiling in Control, hTim8aKO and hTim8a MUT SH cells.
- https://cdn.elifesciences.org/articles/48828/elife-48828-fig5-data1-v3.xlsx
-
Figure 5—source data 2
13C-glucose and 13C-glutamine labelling of Control SH-SY5Y and hTim8MUT SH cells.
- https://cdn.elifesciences.org/articles/48828/elife-48828-fig5-data2-v3.xlsx
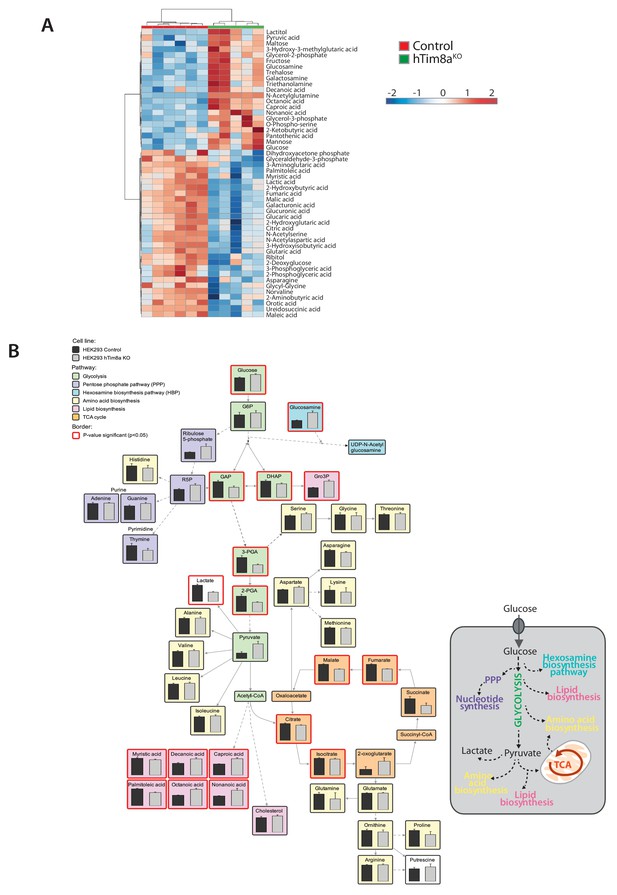
Metabolomic profile in hTim8aKO HEK293 cells.
(A) Hierarchical clustering of relative abundance of the top 40 significantly-affected intracellular metabolites depicted as a heat map (p<0.05) in control (n = 6) and hTim8aKO (n = 5) HEK293 cells. (B) Relative abundance of intracellular metabolites in control and hTim8aKO cells as described in (A) was mapped onto networks using VANTED analysis tool. Pathways are color-coded as indicated and significant metabolites (p<0.05) are boxed using a thicker, red-coloured border. See also Figure 5—source data 1.
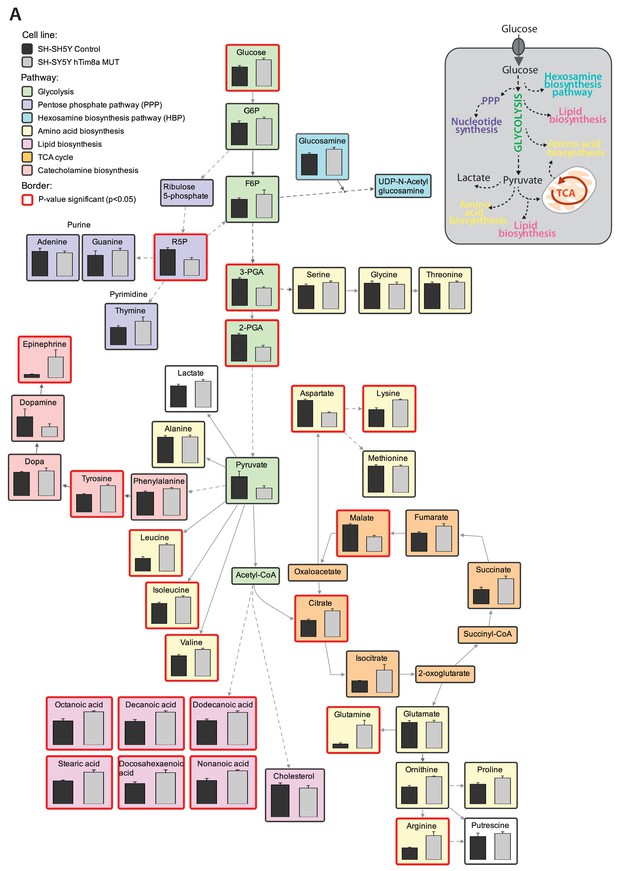
Metabolomic profile in hTim8aMUT SH-SY5Y cells.
(A) Relative abundances of intracellular metabolites in hTim8aMUT compared to control SH-SY5Y cells (n = 6) were mapped onto metabolic networks using VANTED analysis tool. Each pathway is color-coded as indicated and p-value significant metabolites (p<0.05) are boxed using a thicker, red-coloured border.
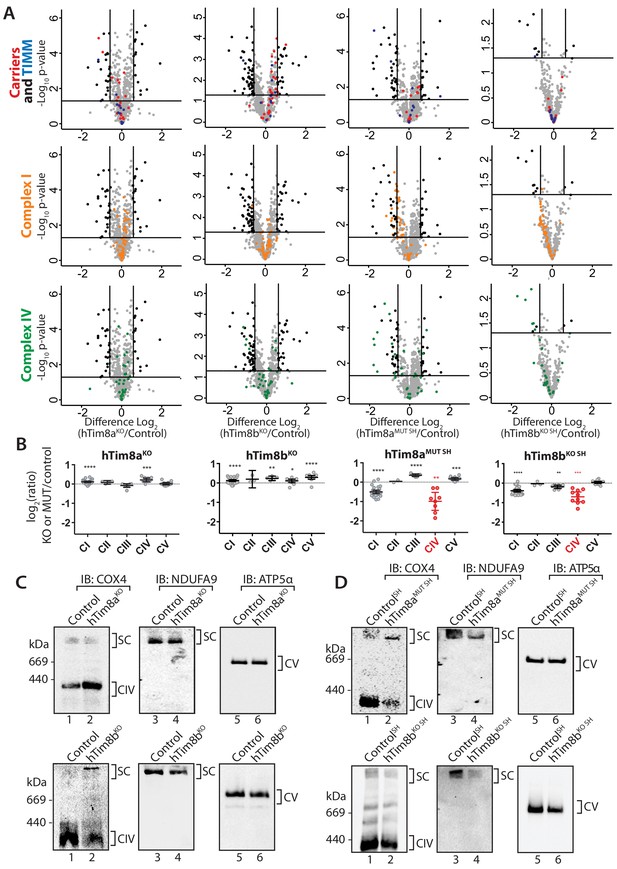
hTim8a and hTim8b have a cell type specific function in Complex IV biogenesis.
(A) Data from label-free quantitative mass spectrometry shown in Figure 2 was used to create volcano plots showing relative levels of proteins in CRISPR-edited cells compared to control cells. n = 3 biological replicates. Significantly altered proteins are located outside the line (p-value<0.05). TIM22 complex subunits/small TIM/hTim23 (blue), carrier proteins of the SLC25 family (red), Complex I subunits (orange) and Complex IV subunits and assembly factors (green) are highlighted and indicated in separate plots. Refer to Figure 2—source data 1. (B) Relative abundance of respiratory chain complexes Complex I to V (subunits and assembly factors) in mitochondria isolated from control and hTim8a andhTim8b CRISPR-edited HEK293 or SH-SY5Y cells, detected using quantitative mass spectrometry in Figure 2 were quantified and tabulated as mean ± SEM (n = 3). *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. (C and D) Mitochondrial lysates from control and hTim8a or hTim8b-CRISPR-edited HEK293 (C) or SH-SY5Y (D) were solubilised in 1% digitonin-containing buffer and analysed using BN-PAGE and immunoblotting with the indicated antibodies to assess for the stability of respiratory chain complexes. See also Figure 6—figure supplement 1.
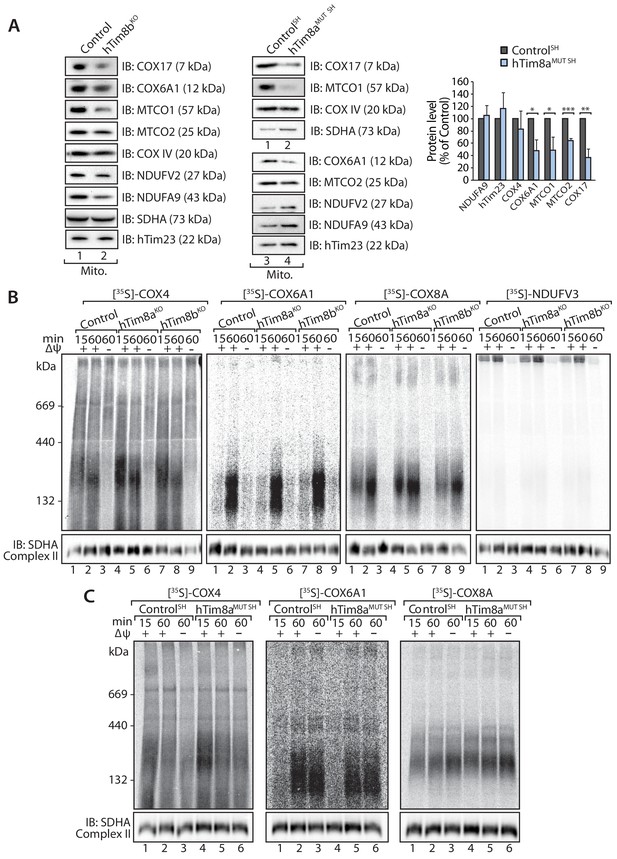
Loss of hTim8a or hTim8b in SH-SY5Y cells leads to altered levels of multiple Complex IV subunits or assembly factors.
(A) Mitochondria isolated from control or hTim8bKO HEK293 or hTim8aMUT SH-SY5Y cells were analysed using SDS-PAGE and immunoblotted with indicated antibodies. Relative abundance of Complex IV subunits and assembly factors were quantified and represented as mean ± SD (n > or = 2). *, p<0.05, **, p<0.01, ***, p<0.001. (B) 35S-labelled Complex IV subunits were imported into mitochondria isolated from control and hTim8aKO or hTim8bKO HEK293 for the indicated time in the presence or absence of membrane potential (ΔΨ) and treated with proteinase K \ prior to solubilisation in 1% digitonin-containing buffer and BN-PAGE analysis. (C) [35S]-labelled Complex IV subunits were imported into mitochondriaisolated from control and hTim8aMUT SH-SY5Y cells for the indicated time in the presence or absence of membrane potential (ΔΨ) and treated with proteinase K prior to solubilisation in 1% digitonin-containing buffer and BN-PAGE analysis.
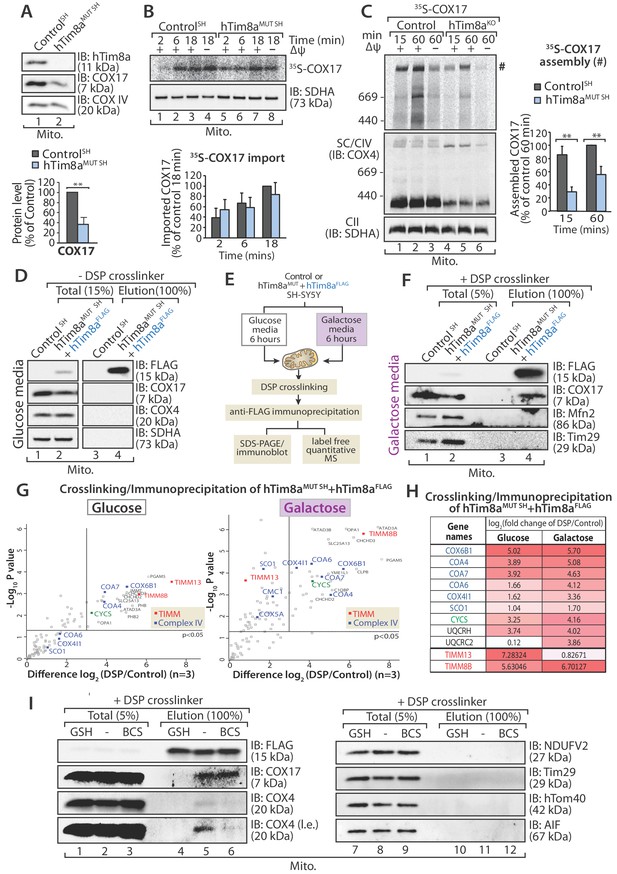
hTim8a functions in the early stage of assembly of Complex IV in SH-SY5Y cells.
(A) Mitochondria isolated from control and hTim8aMUT SH cells were separated using SDS-PAGE and immunoblotted using antibodies against hTim8a, COX17 and COX4. Relative level of COX17 proteins in hTim8aMUT SH-SY5Y mitochondria compared to control were quantified and tabulated as mean ± SD (n = 3). **, p<0.01. (B and C) [35S]-labelled COX17 was incubated with mitochondria isolated from control and hTim8aMUT SH cells in the presence or absence of membrane potential (ΔΨ) and treated with proteinase K and PMSF prior to (B) TCA precipitation for SDS PAGE analysis, or (C) solubilised in 1% digitonin-containing buffer prior to BN-PAGE analysis, and autoradiography. The amount of (B) imported Cox17 (at 18 min) or (C) assembled COX17 (indicated by # on BN-PAGE at 60 min) were quantified and are represented as mean ± SD (n = 2 for (B) SDS-PAGE and n = 3 for (C) BN-PAGE). **, p<0.01. (D) Mitochondria were isolated from control or hTim8aMUT cells re-expressing hTim8aFLAG (hTim8aMUT + hTim8aFLAG) SH-SY5Y cells and solubilised in 0.5% digitonin-containing buffer prior to immunoprecipitation and western blotting using the indicated antibodies. (E) Schematic representation of the assay performed to study the interaction between hTim8a and Complex IV. (F) hTim8aFLAG (hTim8aMUT + hTim8aFLAG) SH-SY5Y were grown in galactose containing media. Mitochondria were isolated and treated for crosslinking with dithiobis-succinimidyl propionate (DSP) and immunoprecipitation with FLAG antibodies. Samples were separated by SDS-PAGE and probed with the indicated antibodies. (G) hTim8aFLAG and control cells were either grown in glucose (left) media or shifted to galactose media for 6 hr (right pabel) prior to mitochondrial isolation and crosslinking with dithiobis-succinimidyl propionate (DSP). Following immunoprecipitation samples were processed for label free quantitative mass spectrometry. n = 3 biological replicates. Significantly altered proteins are located outside the line (p-value<0.05). The small TIM chaperones are coloured in red; and Complex IV subunits or assembly factors are coloured in blue. (H) Table showing the relative fold of enrichment of hTim8a-crosslinked protein partners when cultured in glucose versus galactose media as illustrated in (G): Complex IV subunits/assembly factors (blue), Cytochrome c (CYCS, green), Complex III subunits/assembly factors (black) or hTim8a/13 (red). (I) hTim8aMUT + hTim8aFLAG mitochondria (protein expression induced with doxycycline for 20 hr followed by 6 hr incubation in galactose media) were either left untreated, subjected to bathocuproine disulfonate (BCS) chelation or reduced using glutathione (GSH) prior to DSP crosslinking and immunoprecipitation. Total and eluate fractions were separated using SDS-PAGE and analysed using immunoblotting.
-
Figure 7—source data 1
Interacting partners of hTim8b in SH-SY5Y cells in glucose versus galactose media.
- https://cdn.elifesciences.org/articles/48828/elife-48828-fig7-data1-v3.xlsx
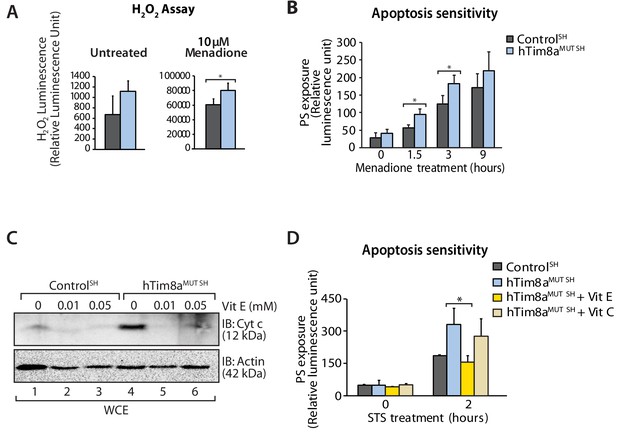
Elevated oxidative stress in hTim8aMUT SH-SY5Y cells sensitises cells to apoptosis.
(A) Reactive H2O2 species present in untreated or menadione-pretreated (2 hr) control and hTim8aMUT SH-SY5Y cells were quantified using ROS-Glo H2O2 Assay. Data are shown as mean ± SD (n = 3, untreated; n = 4, menadione treatment). *, p<0.05. (B) Apoptotic sensitivity of control and hTim8aMUT SH-SY5Y cells was measured following menadione treatment for the indicated time, by assessing phosphatidylserine (PS) exposure (relative luminescence unit). Data is represented as mean ± SD. n = 4 for control and n = 3 for hTim8aMUT SH. *, p<0.05. (C) Mitochondria were isolated from control and hTim8aMUT SH cells following 24 hr of Vitamin E (Vit E, 0, 0.01 or 0.05 mM) treatment prior to SDS-PAGE and immunoblotting analyses. (D) Apoptotic sensitivity of control and hTim8aMUT cells was measured following Vit C (0.2 mM) or Vit E (0.01 mM) treatment by assessing phosphatidylserine (PS) exposure (relative luminescence unit). n = 3, mean ± SD; *, p<0.05.
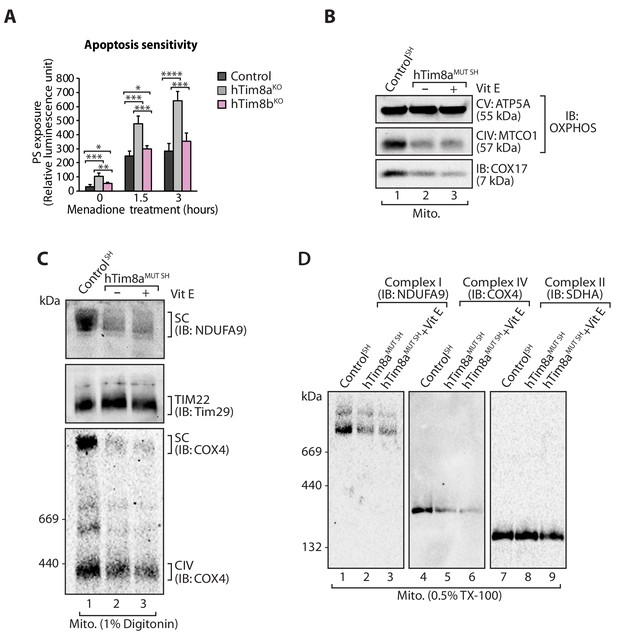
Apoptotic sensitivity in the absence of hTim8a or hTim8b and the effects of Vitamin E treatment.
(A) Control, hTim8aKO and hTim8bKO HEK293 cells were treated with Menadione (10 μM) for 0, 1.5 or 3 hr. Rate of apoptosis was calculated by measuring phosphatidylserine (PS) exposure to the outer leaflet of plasma membrane (relative luminescence unit). n = 3, mean ± SD; *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. (B–D) Mitochondria were isolated from control and hTim8aMUT cells with 24 hr of Vitamin E (Vit E, 0.01 mM) treatment prior to (B) SDS-PAGE or (C) solubilised in 1% digitonin-containing buffer or (D) lysed in 0.5% TX-100-containing buffer, and immunoblotted with the indicated antibodies.
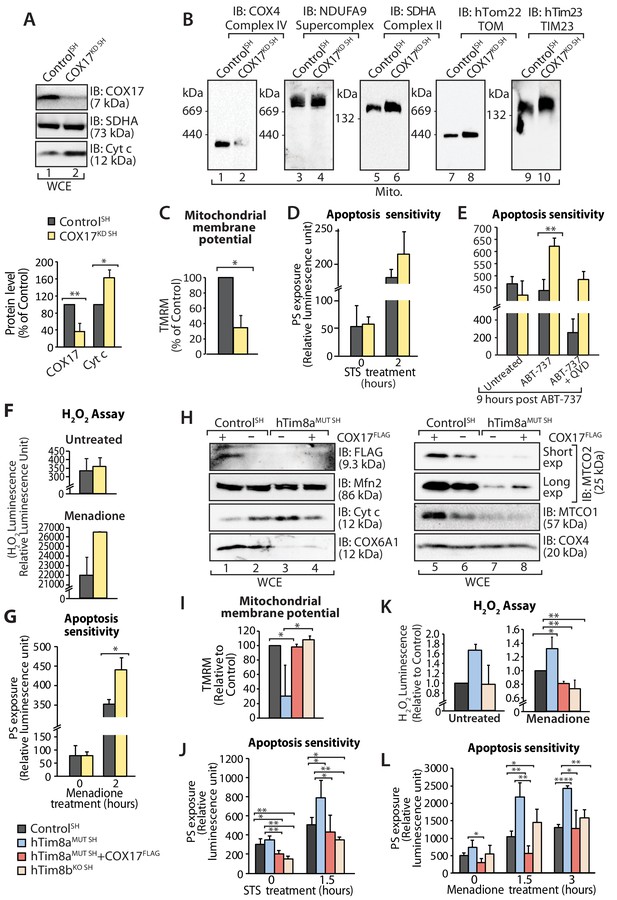
Loss of COX17 in SH-SY5Y cells leads to Complex IV defects and primes cells to apoptotic-cell death.
(A) Cell lysate from SH-SY5Y cells depleted of COX17 (COX17KD SH) using siRNA were analysed using SDS-PAGE and immunoblotted with the indicated antibodies. Relative protein levels of COX17 and cytochrome c (Cyt c) were quantified and tabulated as mean ± SD (n = 3). *, p<0.05, **, p<0.01. (B) Mitochondria isolated from (A) were solubilised in 1% digitonin-containing buffer prior to BN-PAGE/western blot analyses using the indicated antibodies. (C) Control and COX17 siRNA knockdown cells were stained using TMRM and mitochondrial membrane potential was measured and is represented as mean ± SD (n = 4). *, p<0.05. (D) Control and COX17KD SH cells were treated with staurosporine (STS; 1.5 μM) for 0 or 2 hr. Rate of apoptosis was calculated by measuring phosphatidylserine (PS) exposure to the outer leaflet of plasma membrane (relative luminescence unit). n = 3, mean ± SD. (E) Control or COX17KD SH cells were either: (i) left untreated, (ii) treated with ABT-737 (0.1 μM) or (iii) pretreated with QVD-OPh (20 μM) for 20 min prior to ABT-737 treatment, for 9 hr prior to measuring cellular apoptotic sensitivity by assessing phosphatidylserine (PS) exposure (relative luminescence unit). n = 3, mean ± SD. **, p<0.01. (F) Reactive H2O2 species present in untreated or menadione-pretreated (2 hr) control or COX17KD SH-SY5Y cells were quantified using ROS-Glo H2O2 Assay. Data are shown as mean ± SD (n = 4 for untreated, n = 3 for menadione-treatment). (G) Control or COX17KD SH-SY5Y cells were challenged with Menadione for the indicated time prior to measuring cellular apoptotic sensitivity by assessing phosphatidylserine (PS) exposure (relative luminescence unit). n = 3, mean ± SD. (H) Cell lysates from control and hTim8aMUT cells re-expressing COX17FLAG, were analysed using SDS-PAGE and immunoblotted using the indicated antibodies. (I) Control, hTim8aMUT, hTim8aMUT re-expressing COX17FLAG or hTim8bKO SH-SY5Y cells were assessed for mitochondrial membrane potential using TMRM uptake. n = 3, mean ± SD. *, p<0.05. (J) Control, hTim8aMUT, hTim8aMUT re-expressing COX17FLAG or hTim8bKO SH-SY5Y cells were treated with staurosporine (STS, 1.5 μM) for 0 or 1.5 hr prior to measurement of the rate of phosphatidylserine (PS) exposure as an indication of the rate of apoptosis. Data were represented as mean ± SD, n = 4 for control and hTim8aMUT SH re-expressing; n = 3 for hTim8aMUT SH and hTim8bKO SH *, p<0.05; **, p<0.01. (K) Reactive H2O2 species present in untreated or menadione-pretreated (10 μM, 2 hr) control, hTim8aMUT, control re-expressing COX17FLAG, hTim8aMUT re-expressing COX17FLAG or hTim8bKO SH-SY5Y cells were quantified using ROS-Glo H2O2 Assay. Data are shown as mean ± SD (n = 3). *, p<0.05; **, p<0.01. (L) Apoptotic sensitivity of control, hTim8aMUT, hTim8aMUT re-expressing COX17FLAG or hTim8bKO SH-SY5Y cells was measured following menadione (10 μM) treatment for 0, 1.5 or 3 hr, by assessing phosphatidylserine (PS) exposure (relative luminescence unit). n = 3 for control and hTim8aMUT SH re-expressing COX17FLAG; n = 4 hTim8aMUT SH and hTim8bKO SH, mean ± SD; *, p<0.05; **, p<0.01; ****, p<0.0001.





