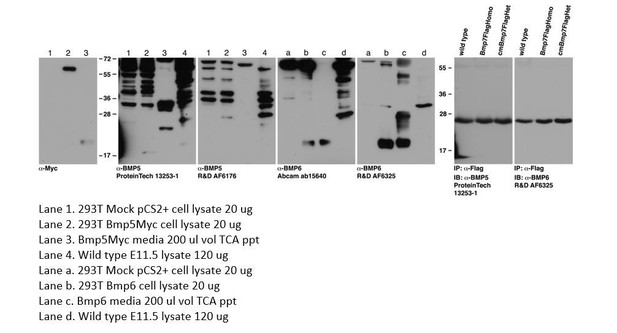
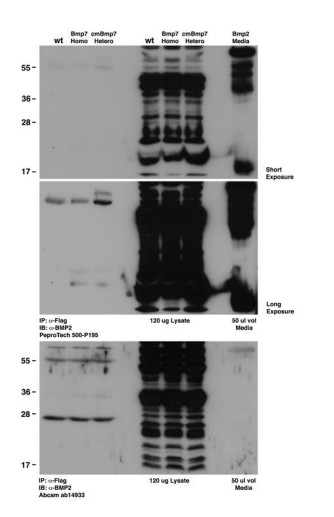
BMP7 functions predominantly as a heterodimer with BMP2 or BMP4 during mammalian embryogenesis
Figures
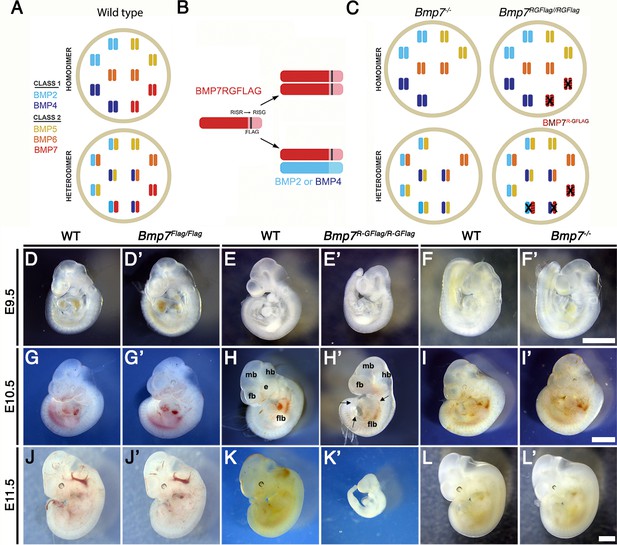
Bmp7R-GFlag homozygotes show earlier lethality and more severe phenotypic defects than Bmp7 null homozygotes.
(A) Illustration of two hypothetical models in which class I and II BMPs function predominantly as homodimers (top) or as heterodimers (bottom) within select cell types. In the heterodimer model, class I/II heterodimers form preferentially. In addition, it is hypothesized that there is an excess of class II BMPs that form homodimers in the wild type condition but are available to form heterodimers to compensate for loss of any single class II BMP. (B) Illustration of BMP7R-GFlag precursor protein forming homodimers (top) or heterodimers with BMP2 or BMP4 (bottom). Prodomain: dark shading, mature domain: light shading, black bar: FLAG epitope. (C) Illustration showing predicted loss of BMP activity in Bmp7R-GFlag or Bmp7 null homozygotes if BMPs function predominantly as homodimers (top) or as heterodimers (bottom). In cells in which only homodimers form, there is predicted to be an equivalent reduction in BMP activity in Bmp7-/- and in Bmp7R-GFlag/Flag mice because only BMP7 homodimers are absent or inactive (black X), respectively. In cells or tissues in which class I/II heterodimers are the primary functional ligand, excess class II molecules that normally form homodimers will fill in to maintain the heterodimer pool in Bmp7 null mutants (lower left), but cannot do so in Bmp7R-GFlag mutants because the BMP7R-GFlag precursor protein forms non-functional covalent heterodimers with endogenous class I BMPs (lower right). This would lead to a greater reduction in the heterodimer pool, lower total BMP activity and more severe phenotypic defects in Bmp7R-GFlag than in then Bmp7 null mutants in any tissues or cell types where heterodimers predominate. (D–L’) Photograph of E9.5–11.5 (age indicated to left of each row) wild type (D–L) or mutant (D’-L’; genotype listed at top of each column) littermates. Scale bars in Panel F’, I’ and L’ correspond to 1 mm and apply across each row. A minimum of eight embryos of each genotype were analyzed at each stage. Arrows in H’ indicate pericardial edema. fb; forebrain, mb; midbrain, hb; hindbrain, flb; forelimb bud, e; eye.
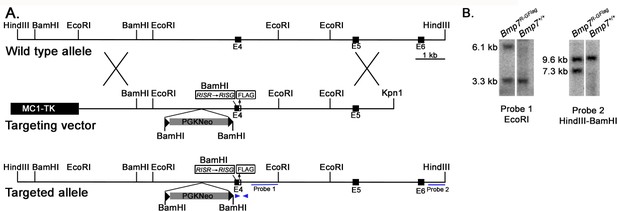
Generation of Bmp7R-GFlag mice.
(A) Genomic organization of the wild type Bmp7 allele, the targeting vector and the Bmp7R-GFlagNeo allele. The positions of the external (probe 2) and internal (probe 1) probes used for Southern analysis, and primers (arrows) surrounding the Flag tag that were used for PCR based genotyping are indicated. (B) Southern blot analysis of genomic DNA from targeted or non-targeted (Bmp7+/+) ES cells. Genomic DNA was digested with EcoRI or HindII and BamHI and hybridized with Probe one or Probe 2.
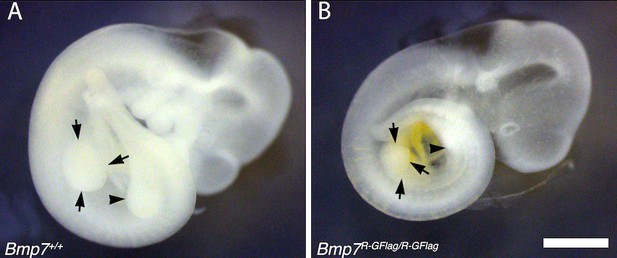
Limb bud size is reduced in Bmp7R-GFlag homozygotes.
E10.5 wild type (A) and Bmp7R-GFlag/R-GFlag (B) littermates are shown. Arrows denote the forelimb bud and arrowhead denotes the hindlimb bud. Scale bar corresponds to 1 mm.
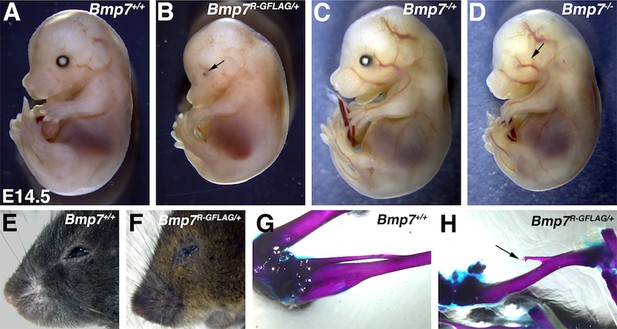
Bmp7R-GFlag heterozygotes show skeletal and eye defects that are absent in Bmp7 null heterozygotes.
(A–F) Photograph of E14.5 (A–D) and adult (E–F) wild type or Bmp7 mutant embryos. Arrows denote small or absent eye in 3 out of 13 Bmp7R-GFlag/+, zero out of eight Bmp7-/+ and five out of five Bmp7-/- mice analyzed between E14.5–18.5. (G–H) Representative skeletal preparations showing that the fibula (arrow) is shortened and not attached to the knee in two out of fourteen adult (3–5 months old) Bmp7R-GFlag heterozygotes that were examined.
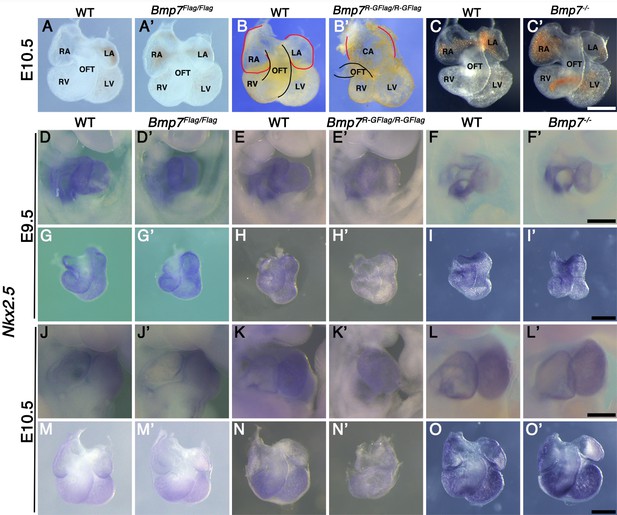
Bmp7R-GFlag homozygotes show defects in heart development that are absent in Bmp7 null homozygotes.
(A–C’) Photographs of hearts dissected from E10.5 mice carrying targeted alleles of Bmp7 (A’–C’) and wild type littermates (A–C). Genotypes are indicated above each panel. The OFT is outlined in black, and the atrium in red in B and B’. (D–O’) Expression of Nkx2.5 was analyzed by whole mount in situ hybridization in mice carrying targeted alleles of Bmp7 (D’–O’) and wild type littermates (D–O) at E9.5 or E10.5 as indicated to the left of each row. Genotypes are indicated above each panel. Close up photographs of hearts in intact embryos (D–F’, J–L’) or dissected free of embryos (G–I’, M–O’) are shown. RA; right atrium, LA; left atrium, CA; common atrium, RV; right ventricle, LV; left ventricle, OFT; outflow tract. Scale bars in all panels correspond to 500 µM.
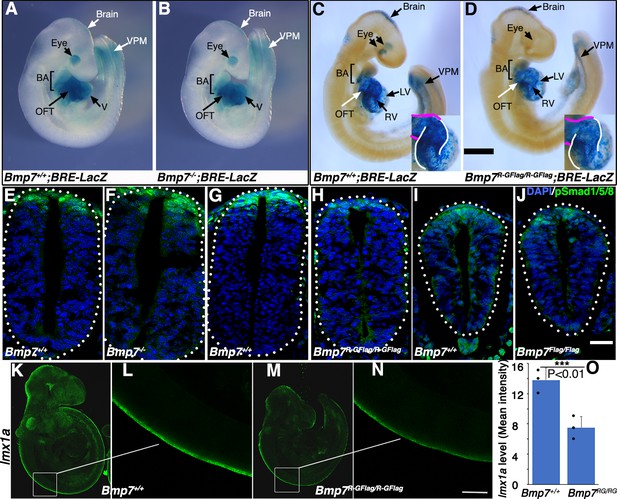
Bmp7R-GFlag mutants, but not Bmp7 null mutants, show reduced BMP activity in multiple tissues.
(A–D) E9.5 Bmp7-/- (B), Bmp7R-GFlag/R-GFlag (D) and wild type littermates (A, C) carrying a BRE-LacZ transgene were stained for ß-galactosidase activity to detect endogenous BMP pathway activation. Embryos from a single litter were stained for an identical period of time under identical conditions. A minimum of three embryos of each genotype were examined and results shown were reproduced in all. VPM; ventral posterior mesoderm, BA; branchial arches, OFT; outflow tract, RV; right ventricle, LV; left ventricle V; ventricles. Insets in C, D show an enlarged view of the heart with the OFT and RV outlined in white and magenta, respectively. In C, left and right eyes are visible in the cleared embryo. Scale bars correspond to 1 mm. (E–H) Transverse sections through the spinal cord of E9.5 Bmp7-/- (F), Bmp7R-GFlag/R-GFlag (H), Bmp7Flag/Flag (J), and wild type littermates (E, G, I) were immunostained with antibodies specific for pSmad1/5/8 and nuclei were stained with DAPI. Dorsal is up. Scale bars correspond to 20 µm. Three embryos of each genotype were analyzed and results were reproduced in all. (K–N) In situ HCR was used to analyze expression of lmx1a in three E9.5 Bmp7R-GFlag/R-GFlag (M–N) and wild type littermates (K–L). White boxes in K and M indicate the region of staining shown in L and N. Scale bar corresponds to 100 µm. (O) Levels of lmxa1 transcripts (mean fluorescence ± s.d., data analyzed by two tailed t-test). Fluorescence intensity was quantified in comparable regions of the spinal cord in three embryos of each genotype. Dots on the graph denote values for individual embryos.
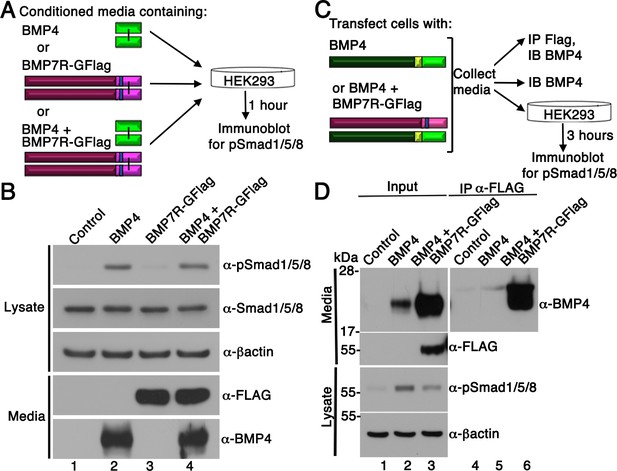
BMP7R-GFlag homodimers form inside of cells and cannot act outside of cells to block BMP signaling.
(A) HEK293T cells were cultured for one hour in conditioned media containing equivalent amounts of BMP4 ligand alone, BMP7R-GFlag precursor protein alone or BMP4 ligand and BMP7R-GFlag precursor together as illustrated. (B) Proteins were separated by electrophoresis under reducing conditions and levels of pSmad1/5/8, total Smad1/5/8, BMP7R-GFlag and BMP4 (monomers) were analyzed by immunoblot. Blots were reprobed for ß-actin as a loading control. Results were reproduced in three independent experiments. (C, D) HEK293T cells were transfected with 700 ng vector, 200 ng BMP4 + 500 ng vector or 200 ng BMP4 + 500 ng BMP7R-GFlag. Equivalent volumes of conditioned media were subjected to trichloracetic acid precipitation or to immunoprecipitation (IP) with anti-Flag antibodies. Proteins were separated by electrophoresis under reducing conditions prior to immunoblotting (IB) with antibodies specific for BMP4 or Flag to detect BMP4 and BMP7 monomers. HEK293T cells were cultured for three hours in equivalent volumes of conditioned media from cells transfected with vector, BMP4 alone, or BMP4 + BMP7R-GFlag. Levels of pSmad1/5/8 were analyzed by immunoblot. Blots were reprobed for ß-actin as a loading control. Results were reproduced in two independent experiments.
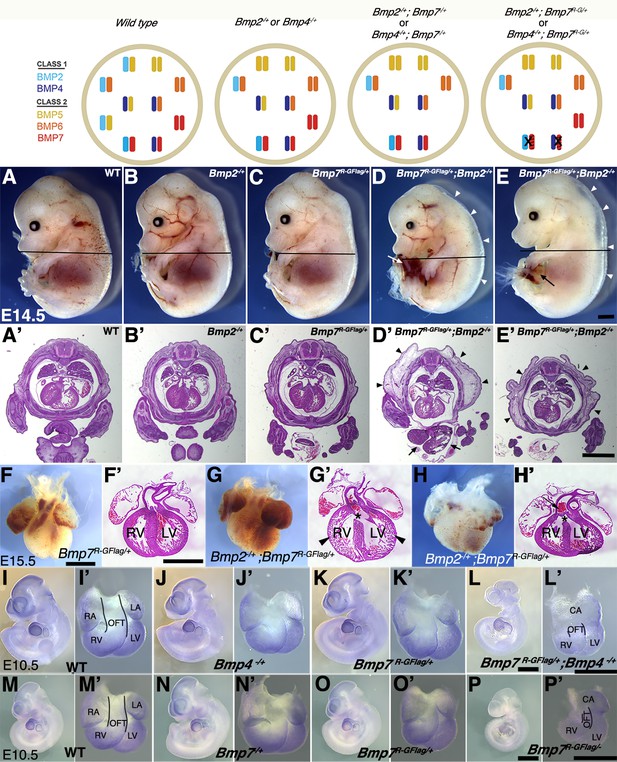
Analysis of compound heterozygotes shows that BMP2/7 and BMP4/7 heterodimers are required for early development.
Schematic illustration of BMP activity in embryos carrying wild type or mutant alleles of Bmp2, 4 and/or 7. In the hypothetical model, class I/II heterodimers form preferentially. In addition, it is hypothesized that there is an excess of class II BMPs that form homodimers in the wild type condition but are available to form heterodimers to compensate for loss of any single class II BMP. In embryos lacking a single copy of Bmp2 or Bmp4, activity contributed by the heterodimer pool is reduced but there is no further reduction in heterodimers when a single copy of Bmp7 is also removed, due to redundancy with other Class II BMPs. A fraction of the heterodimer pool is inactivated in Bmp7R-GFlag heterozygotes and additional removal of a single copy of Bmp2 or Bmp4 further depletes the heterodimer pool. (A–E’) Photographs of E14.5 wild type and mutant littermates (A–E) and corresponding hematoxylin and eosin stained transverse sections through the abdomen (A’-E’; approximate position of section indicated by the black bar in A-E). Arrows indicated externalized viscera (D, D’, E) and arrowheads denote peripheral edema (D, D’, E, E’). (F–H’) Ventral views of hearts dissected from E15.5 Bmp7R-GFlag/+ (F) or Bmp7R-GFlag/+;Bmp2-/+ embryos (G–H) and corresponding hematoxylin and eosin stained coronal sections (F’–H’). Asterisks denote VSDs (G’, H’), arrow indicates abnormal positioning of the aorta exiting the right ventricle (H’), arrowheads indicate thin, non-compacted ventricular wall (G’). (I–P’) Expression of Nkx2.5 was analyzed by whole mount in situ hybridization in littermates generated by intercrossing Bmp4-/+ and Bmp7R-GFlag/+ (I–L’) or Bmp7-/+ and Bmp7R-GFlag/+ mice (M–P’). Photographs of intact embryos at E10.5 (I–P) and photographs of hearts dissected from corresponding embryos (I’–P’) are shown. RA; right atrium, LA; left atrium, CA; common atrium, RV; right ventricle, LV; left ventricle, OFT; outflow tract. The OFT is outlined in I’, L’, M’ and P’. Scale bars in all panels correspond to 1 mm.
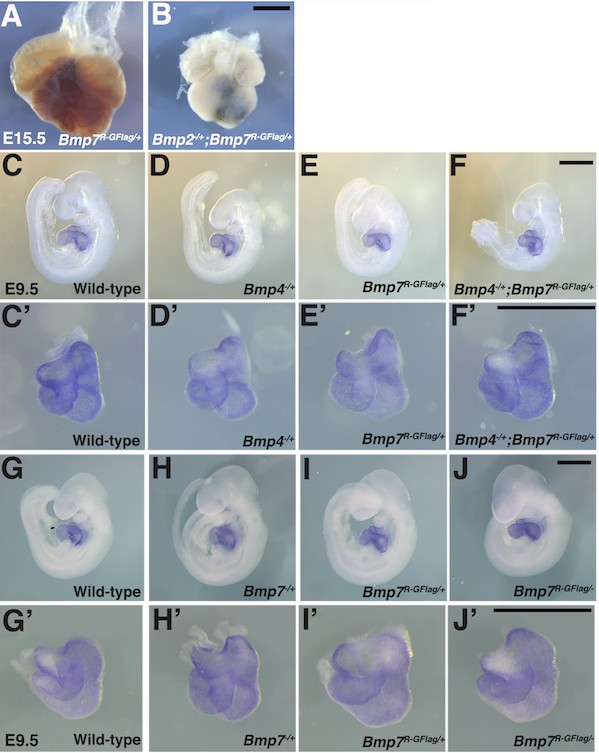
Heart defects are present in Bmp2-/+;Bmp7R-GFlag/+ embryos at E15.5 and are absent in Bmp4-/+;Bmp7R-GFlag/+ and Bmp7R-GFlag/- embryos at E9.5.
(A–B) Photograph of heart dissected from Bmp2-/+;Bmp7R-GFlag/+ embryo and Bmp7R-GFlag/+ littermate. (C–J’) Expression of Nkx2.5 was analyzed by whole mount in situ hybridization in littermates generated by intercrossing Bmp4-/+ and Bmp7R-GFlag/+ (C–F’) or Bmp7-/+ and Bmp7R-GFlag/+ mice (G–J’). Photographs of intact embryos at E9.5 (C–J) and photographs of hearts dissected from corresponding embryos (C’–J’) are shown. Scale bars in all panels correspond to 1 mm.
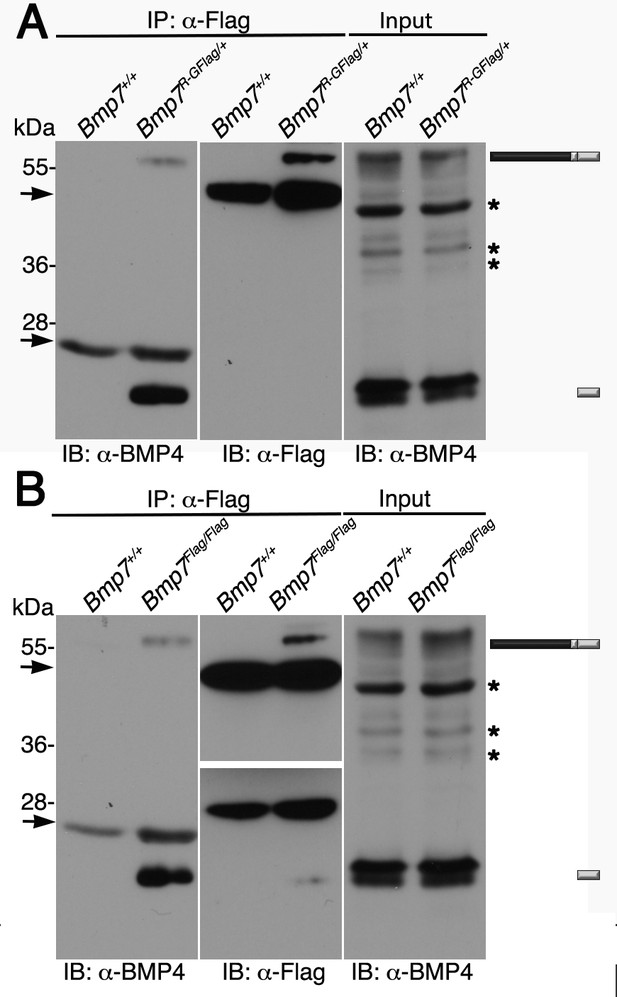
Endogenous BMP4 co-immunoprecipitates with BMP7.
(A) Antibodies specific for the Flag-epitope tag were used to immunoprecipitate (IP) proteins from E11.5 Bmp7+/+ or Bmp7R-GFlag/+ lysates. Immunoblots (IB) of IPs or total protein (input) were probed with antibodies specific for BMP4 or Flag as indicated below each panel. (B) Antibodies specific for the Flag-epitope tag were used to immunoprecipitate (IP) proteins from E11.5 Bmp7+/+ or Bmp7Flag/Flag lysates. Immunoblots (IB) of IPs or total protein (input) were probed with antibodies specific for BMP4 or Flag as indicated below each panel. (A, B) The position of precursor proteins and cleaved mature ligand is indicated on the right. Arrows denote bands corresponding to IgG heavy or light chains. The band at 28 kDa on the middle panel is a non-specific band detected by HRP-conjugated Flag antibody. Asterisks mark bands that are considered non-specific because only the bands marked as precursor and ligand show decreased intensity on BMP4 immunoblots of lysates from Bmp4+/- relative to Bmp4+/+ embryos (Tilak et al., 2014) and unpublished results). Results were reproduced in three independent experiments.
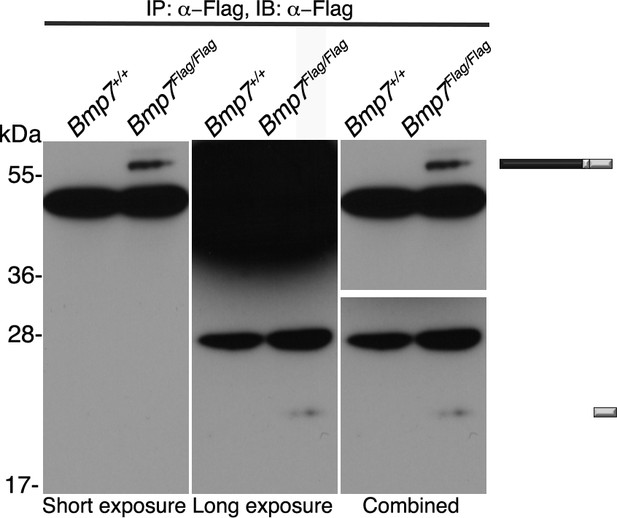
Short and long exposure of Flag immunoblot.
Antibodies specific for the Flag-epitope tag were used to immunoprecipitate (IP) proteins from E11.5 Bmp7+/+ or Bmp7Flag/Flag lysates. Immunoblots IPs were probed with antibodies specific for Flag. Middle and right panels are short and long exposures, respectively, of the same blot. The position of precursor proteins and cleaved mature ligand is indicated on the right.
Tables
Progeny from Bmp7Flag/+ intercrosses
https://doi.org/10.7554/eLife.48872.006| Age | Bmp7+/+ | Bmp7Flag/+ | Bmp7Flag/Flag | Total |
|---|---|---|---|---|
| P28 | 8 (20%) | 24 (60%) | 8 (20%) | 40 |
| E9.5-14.5 | 23 (31%) | 34 (46%) | 17 (23%) | 74 |
-
Data are presented as number (percent).
Progeny from Bmp7-/+ intercrosses
https://doi.org/10.7554/eLife.48872.007| Age | Bmp7+/+ | Bmp7-/+ | Bmp7-/- | Total |
|---|---|---|---|---|
| E13.5-18.5 | 12 (30%) | 18 (45%) | 10 (25%) | 40 |
| E9-12.5 | 21 (32%) | 32 (48%) | 13 (20%) | 66 |
-
Data are presented as number (percent).
Progeny from Bmp7R-GFlag/+ intercrosses
https://doi.org/10.7554/eLife.48872.008| Age | Bmp7+/+ | Bmp7R-GFlag/+ | Bmp7R-GFlag/R-GFlag | Total |
|---|---|---|---|---|
| E18.5-P0** | 15 (52%) | 14 (41%) | 0 (0%) | 29 |
| E12.5-E14.5* | 9 (47%) | 11 (53%) | 0 (0%) | 20 |
| E11.5 | 12 (29%) | 18(42%) | 12 (29%) | 42 |
| E10.5 | 6 (17%) | 16 (46%) | 13 (37%) | 35 |
| E9.5 | 28 (18%) | 83 (53%) | 45 (29%) | 156 |
-
Data are presented as number (percent). Asterisks indicate that the observed frequency is significantly different than the expected frequency by Chi-square analysis (**P<0.01).
Progeny from Bmp7R-GFlag/+ and Bmp2-/+ intercrosses
https://doi.org/10.7554/eLife.48872.014| Age | Wildtype | Bmp2-/+ | Bmp7R-GFlag/+ | Bmp2-/+;Bmp7R-GFlag/+ | Total |
|---|---|---|---|---|---|
| P21** | 12 (50%) | 7 (29%) | 5 (29%) | 0 (0%) | 24 |
| E15.5-16.5 | 8 (26%) | 8 (26%) | 10 (32%) | 5 (16%) | 31 |
| E14.5 | 18 (37%) | 11 (23%) | 10 (20%) | 10 (20%) | 49 |
-
Data are presented as number (percent). Asterisks indicate that the observed frequency is significantly different than the expected frequency by Chi-square analysis (**P<0.01, *P<0.05).
Progeny from Bmp7R-GFlag/+ and Bmp4-/+ intercrosses
https://doi.org/10.7554/eLife.48872.015| Age | Wildtype | Bmp4-/+ | Bmp7R-GFlag/+ | Bmp4-/+;Bmp7R-GFlag/+ | Total |
|---|---|---|---|---|---|
| E12.5-14.5* | 9 (38%) | 8 (33%) | 7 (29%) | 0 (0%) | 24 |
| E11.5 | 5 (24%) | 4 (19%) | 5 (24%) | 7 (33%) | 21 |
| E10.5 | 5 (16%) | 7 (23%) | 11 (35%) | 8 (26%) | 31 |
| E9.5 | 14 (34%) | 11 (27%) | 7 (17%) | 9 (22%) | 41 |
-
Data are presented as number (percent). Asterisks indicate that the observed frequency is significantly different than the expected frequency by Chi-square analysis (*P<0.05).
Progeny from Bmp7R-GFlag/+ and Bmp7-/+ intercrosses
https://doi.org/10.7554/eLife.48872.016| Age | Wildtype | Bmp7-/+ | Bmp7R-GFlag/+ | Bmp7R-GFlag/- | Total |
|---|---|---|---|---|---|
| E12.5-14.5* | 7 (29%) | 9 (38%) | 8 (33%) | 0 (%) | 24 |
| E10.5 | 9 (37%) | 5 (21%) | 6 (25%) | 4 (17%) | 24 |
| E9.5 | 4 (17%) | 7 (31%) | 6 (26%) | 6 (26%) | 23 |
-
Data are presented as number (percent). Asterisks indicate that the observed frequency is significantly different than the expected frequency by Chi-square analysis (*P<0.05).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Bmp4-/+ | PMID: 10049358 | RRID:MGI:2664348 | Dr. Brigid Hogan (Duke University) |
| Genetic reagent (M. musculus) | BRE-LacZ | PMID: 15331632 | Dr. Christine Mummery (Leiden University) | |
| Genetic reagent (M. musculus) | Bmp2-/+ | PMID: 8898212 | RRID:MGI:2658703 | Dr. Yuji Mishina (University of Michigan) |
| Genetic reagent (M. musculus) | Bmp7-/+ | PMID: 9693150 | RRID:IMSR_EM:02513 | Dr. Elizabeth Robertson (University of Oxford) |
| Genetic reagent (M. musculus) | Bmp7flox/flox | PMID: 22219353 | RRID:MGI:5312875 | Dr. James Martin (Baylor University) |
| Genetic reagent (M. musculus) | CMV-CRE | PMID: 8559668 | RRID:IMSR_JAX:006054 | University of Utah, Transgenic Mouse Facility |
| Genetic reagent (M. musculus) | Bmp7R-GFlag | This paper | Generated using gene targeting technology | |
| Genetic reagent (M. musculus) | Bmp7Flag | This paper | Generated using CRISPR-Cas9 technology | |
| Transfected construct (Synthesized) | CS2+BMP4 | PMID: 15356272 | Catherine Degnin (Oregon Health and Science University) | |
| Transfected construct (Synthesized) | CS2+BMP7R-GFlag | This paper | PCR used to insert Flag tag in cDNA | |
| Antibody | Rabbit polyclonal anti-pSmad1/5/8 | Cell Signaling | Cat. #9511S | WB (1:1000), IHC (1:500) |
| Antibody | Rabbit polyclonal anti-Alexa Fluor 488 | Invitrogen | Cat. #11008 | IHC (1:500) |
| Antibody | Mouse monoclonal anti-BMP4 | Santa Cruz | Cat. # sc-12721 | WB (1:1000) |
| Antibody | Mouse monoclonal anti-Flag M2 | Sigma | Cat. # F1804 | WB (1:1000) |
| Antibody | HRP-conjugated mouse monoclonal Flag M2 | Sigma | Cat. # A8592 | WB (1:5,000) |
| Antibody | Rabbit polyclonal anti-beta actin | AbCam | Cat. # ab8227 | WB (1:10,000) |
| Antibody | HRP-conjugated anti-rabbit polyclonal IgG | Jackson ImmunoResearch | Cat. # 111-035-144 | WB (1:10,000) |
| Antibody | HRP-conjugated anti-mouse polyclonal IgG2b | Jackson ImmunoResearch | Cat. # 115-035-207 | WB (1:10,000) |
| Antibody | Anti-Flag mouse monoclonal M2 Agarose | Sigma | Cat. # A2220 | IP (1:500) |
| Cell line (H. sapiens) | HEK293T | American Type Culture Collection | Cat. # CRL-11268, RRID:CVCL_0045 | |
| Commercial assay or kit | lmx1A probe set, HCR amplification and buffer | Molecular Instruments | ||
| Commercial assay or kit | ECL Prime Western Kit | Fisher | Cat. # 45-010-090 | |
| Commercial assay or kit | BCA Protein Assay Kit | Fisher | Cat. # 23225 | |
| chemical compound, drug | Halt protein and phosphatase inhibitor | Fisher | Cat. # 78442 |
Additional files
-
Supplementary file 1
Progeny from Bmp7R-GFlag/+ and Bmp7+/+ intercrosses.
Numbers and percent of animal of each genotype at P28.
- https://doi.org/10.7554/eLife.48872.019
-
Supplementary file 2
Progeny from Bmp7Flag/+ and Bmp2-/+, Bmp4-/+, and Bmp7-/+ intercrosses.
(A) Progeny from Bmp7Flag/+ and Bmp2-/+ intercrosses. (B) Progeny from Bmp7Flag/+ and Bmp4-/+ intercrosses. (C) Progeny from Bmp7Flag/+ and Bmp7-/+ intercrosses.
- https://doi.org/10.7554/eLife.48872.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.48872.021