Gene Drive: A family of killers
Some genes are born criminals. Mendel’s law of segregation states that the two alleles of a heterozygote are each transmitted into half of the gametes, but genes called meiotic drivers break this law by striving to be the only gene variant inherited. This selfish behavior can help them to spread in a population, but it can also decrease the fitness of the organisms carrying the loci (Sandler and Novitski, 1957; Zimmering et al., 1970; Zanders and Unckless, 2019).
Meiotic drivers employ a variety of self-promoting mechanisms (Lindholm et al., 2016). Some are particularly ruthless because they kill the gametes or spores that do not inherit the drive gene. Meiotic drivers are found throughout eukaryotes, including mammals, but relatively little is known about the genes responsible. Now, in eLife, Hanna Johannesson and co-workers at Uppsala University, the University of Bordeaux and Wageningen University – including Aaron Vogan and Lorena Ament-Velásquez, both from Uppsala, as joint first authors – report on the genetic architecture underlying a series of killer meiotic drive phenotypes in the fungus Podospora anserina (Vogan et al., 2019).
P. anserina is an excellent model organism for studying meiotic drive, largely because the phenotype is easy to observe: the two spores that inherit the drive locus are viable and pigmented, while the two spores that do not are dead and unpigmented (Figure 1A; Padieu and Bernet, 1967; Turner and Perkins, 1991; Espagne et al., 2008; Dalstra et al., 2003).
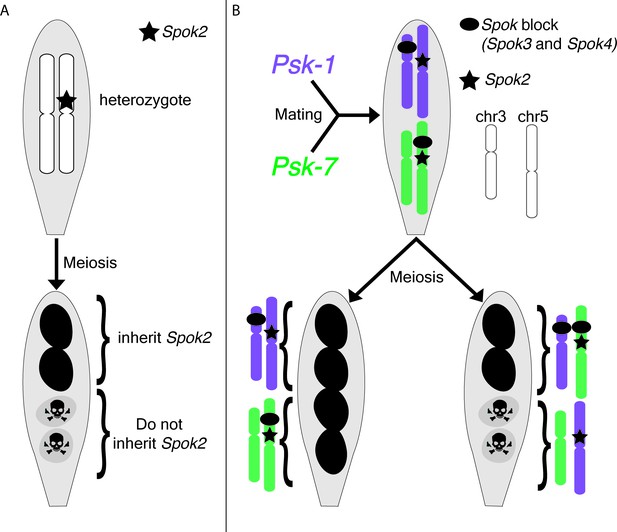
Meiotic drive in Podospora anserina.
(A) A heterozygous diploid generated by mating a naïve strain to one carrying Spok2 undergoes meiosis and generates four spores. One possible outcome of meiosis is depicted. The spores that inherit Spok2 are alive (pigmented), while the spores that do not inherit the Spok2 gene are destroyed (unpigmented, hazard signs). (B) A heterozygous diploid generated by mating the Psk-1 strain (purple) to the Psk-7 strain (green) undergoes meiosis and generates four spores. Note that the Spok block (black oval) is on different chromosomes in the two isolates. Two possible outcomes are depicted: (left) all four spores survive because they all inherited Spok2 and the block that contains Spok3 and Spok4; (right) two of the four spores do not survive because they did not inherit the Spok block.
P. anserina strains carrying spore-killing loci have been sorted into seven types (Psks) based on their killing phenotypes, but the genes underlying these phenotypes were unknown (van der Gaag et al., 2000). Recently, Spok2 was identified as an autonomous single-gene meiotic driver in P. anserina, and it was shown that the Spok genes comprise a gene family found in many copies in diverse fungal lineages (Grognet et al., 2014). These discoveries suggested the Spok genes as candidates for causing the Psk phenotypes.
Vogan et al. started by sequencing and assembling the genomes of six P. anserina strains representing the distinct spore-killing phenotypes. These assemblies showed that Spok2 is present in most P. anserina isolates and facilitated the discovery of two new Spok genes (Spok3 and Spok4) that are both spore killers. Curiously, these two genes are found within a block of sequence that is found in single copy at different locations within the various genomes. In some, the block contains only Spok3 or Spok4, but in others it contains both. How the block has moved during the evolutionary history of P. anserina is a mystery.
Next, Vogan et al. complemented their genomic approach with classical genetic analyses. They demonstrated that all the Psk phenotypes can be explained by just three genes (Spok2, Spok3 and Spok4). Spore killing occurs when two out of the four spores fail to inherit one or more of the Spok genes present in the diploid cell that underwent meiosis (Figure 1A and B). Strains with no Spok genes are considered naïve and spore killing will occur when these strains are crossed to Psk strains, but not to other naïve strains. The most dominant Psk phenotypes, Psk-1 and Psk-7, contain all three Spok genes. These strains cause killing when crossed to all other types. Interestingly, killing is also observed when Psk-1 and Psk-7 are crossed to each other because the Spok block is found at different loci in the two strains, which means that some spores do not inherit it (Figure 1B). The other strains that cause killing contain one or two Spok genes. Again, their phenotypes are explained by the identity and location of their Spok genes.
The latest work also provides a significant advance in our understanding of the mechanisms used by Spok genes. Previous work demonstrated that these genes encode both killing and resistance functions in genetically separable domains and predicted a kinase domain in one of these regions (Grognet et al., 2014). The new work shows that Spok3 can be expressed outside of spore formation, and Vogan et al. exploit this observation to better characterize how single Spok genes carry out their dual roles. In particular they show that the region required for the resistance function must include a cysteine-rich domain (in addition to the kinase domain mentioned above). Moreover, they identify a nuclease domain and demonstrate that it is required for the killing function. However, how the nuclease domain promotes killing and how the kinase domain neutralizes that killing are unknown.
It is becoming clear that genetic criminals that break Mendel’s law of segregation are not rare. Discovering the mechanisms used by meiotic drive genes and how they evolve is necessary to fully understand gametogenesis. Exciting discoveries in Podospora will continue to shape our understanding of the molecular genetics of meiotic drive.
References
-
Genes that bias Mendelian segregationPLOS Genetics 10:e1004387.https://doi.org/10.1371/journal.pgen.1004387
-
The ecology and evolutionary dynamics of meiotic driveTrends in Ecology & Evolution 31:315–326.https://doi.org/10.1016/j.tree.2016.02.001
-
[Mode of action of the genes responsible for abortion of certain products of meiosis in the ascomycete, Podospora anserina]Comptes Rendus Hebdomadaires Des Seances De l'Academie Des Sciences. Serie D: Sciences Naturelles 264:2300–2303.
-
Meiotic drive as an evolutionary forceThe American Naturalist 91:105–110.https://doi.org/10.1086/281969
-
Meiotic drive in Neurospora and other fungiThe American Naturalist 137:416–429.https://doi.org/10.1086/285174
-
Spore-killing meiotic drive factors in a natural population of the fungus Podospora anserinaGenetics 156:593–605.
-
Fertility costs of meiotic driversCurrent Biology 29:R512–R520.https://doi.org/10.1016/j.cub.2019.03.046
-
Mechanisms of meiotic driveAnnual Review of Genetics 4:409–436.https://doi.org/10.1146/annurev.ge.04.120170.002205
Article and author information
Author details
Publication history
Copyright
© 2019, De Carvalho and Zanders
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,231
- views
-
- 111
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.






