A potent voltage-gated calcium channel inhibitor engineered from a nanobody targeted to auxiliary CaVβ subunits
Figures
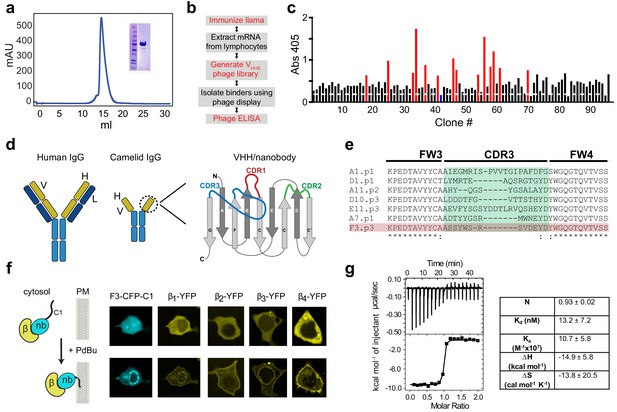
Development of a pan-CaVβ nanobody.
(a) Size-exclusion chromatograph and Coomassie gel (inset) showing purified Cavβ1 from baculovirus-infected HEK293 GnTl- cells. (b) Flow-chart of nanobody generation. (c) Phage ELISA using CaVβ1 as bait and periplasmic extracts from single infected E. coli clones. Red bars represent clones that were selected for subsequent analyses; blue bar represents a negative control from an E. coli expressing an anti-GFP nanobody. (d) Cartoon showing conventional IgG antibody (left) and camelid heavy-chain antibody (center). Right, a schematic representation of the variable heavy chain (VHH or nanobody) of camelid heavy-chain antibodies. The three CDR loops which are the primary determinants of antigen-binding are shown in red, green, and blue. (e) Sequence alignment of CDR3 from selected clones. (f) Left, schematic of co-translocation assay to determine nanobody/CaVβ interaction in HEK293 cells. Right, confocal images showing membrane co-translocation of CaVβX-YFP and nb.F3-CFP-C1PKCγ in response to treatment with 1 uM phorbol 12,13-dibutyrate (PdBu). (g) Left, exemplar isothermal titration calorimetry trace using purified CaVβ2b and nb.F3. Right, summary of ITC thermodynamic parameters. N, stoichiometry; Kd, dissociation constant; Ka, affinity constant; ∆H, enthalpic change; ∆S entropic change. T = 298K.
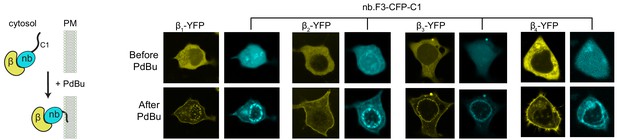
Nb.F3 binds all four CaVβ subunits in the cytosol of mammalian cells Left, schematic of phorbol ester 12,13-dibutyrate (PdBu) translocation assay.
Right, confocal images of HEK293 cells expressing nb.F3-CFP-C1PKCγ and a YFP-CaVβ before (top) and after (bottom) the addition of 1 µM Pdbu.
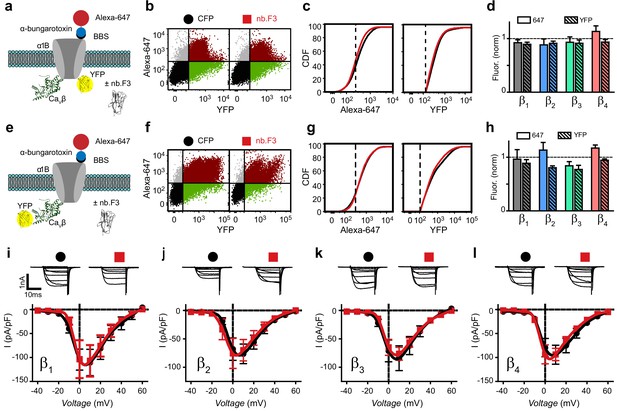
Nb.F3 is functionally silent on reconstituted CaV2.2 channels.
(a) Schematic of experimental strategy; BBS-α1B-YFP was transfected in HEK293 cells with α2δ, CaVβ and either CFP or nb.F3-P2A-CFP. (b) Exemplar flow cytometry dot plot of cells expressing BBS-α1B-YFP + CaVβ1 + α2δ-1 and either CFP (left) or nb.F3-P2A-CFP (right). Approximately 100,000 cells are represented here and throughout. Horizontal and vertical lines represent the threshold for YFP- and Alexa-647-positive cells, respectively, as determined with single color controls. (c) Cumulative distribution histogram of Alexa-647 (left) or YFP fluorescence (right) from CFP (black) or nb.F3 (red) expressing cells. YFP-positive cells were selected for the analysis; dashed lines represent thresholds for Alexa-647 and YFP fluorescence signals above background. (d) Summary flow cytometry data of surface (647, filled) and total (YFP, patterned) levels of BBS-α1B-YFP. Data from nb.F3-expressing cells was normalized to CFP control group. n=>5,000 cells analyzed per experiment, N=4 separate experiments, error bars, s.e.m. (e) Experimental strategy; HEK293 cells were transfected with BBS-α1B + CaVβ-YFP + α2δ-1. (f-h) Same format as (b-d) for cells expressing BBS-α1B + CaVβ-YFP+ α2δ-1 ± nb.F3-P2A-CFP. (i) Exemplar whole-cell Ba2+ currents (top) and population I-V curves (bottom) in HEK293 cells expressing α1B + CaVβ1 + α2δ-1 and either CFP (black) or nb.F3-P2A-CFP (red). (j-l) Same format as (i) for cells expressing CaVβ2 (j), CaVβ3 (k), and CaVβ4 (l). Scale bar 1 nA, 10 ms. Data are means ± s.e.m., n=10 for each point.
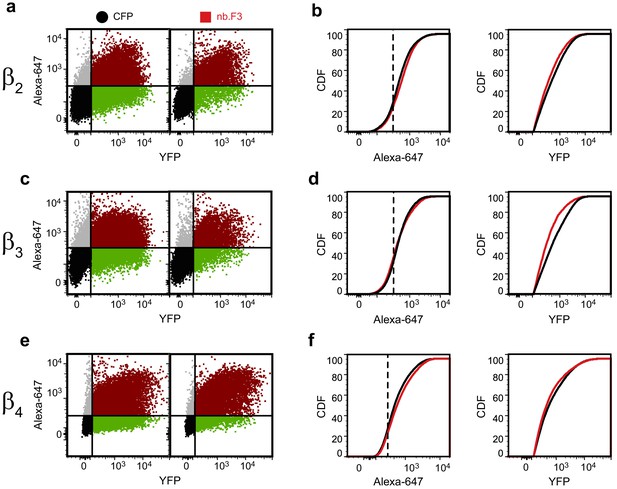
Exemplar flow cytometry data for BBS-α1B with YFP-CaVβ2- CaVβ4.
(a) Exemplar flow cytometry dot plot of cells transfected with BBS-α1B, α2-δ, YFP-CaVβ2 and CFP (left) or nb.F3 (right). (b) Cumulative distribution histogram of data from (a) of Alexa-647 (left) or YFP fluorescence (right) from CFP (black) or nb.F3 (red) expressing cells. YFP-positive cells (n => 5,000 cells/experiment) were selected for analysis; the threshold for 647 labeling and YFP fluorescence above background is represented with the dashed line. (c,d) as in (a,b) for cells expressing YFP-CaVβ3. (e,f) as in (a,b) for cells expressing CaVβ4..
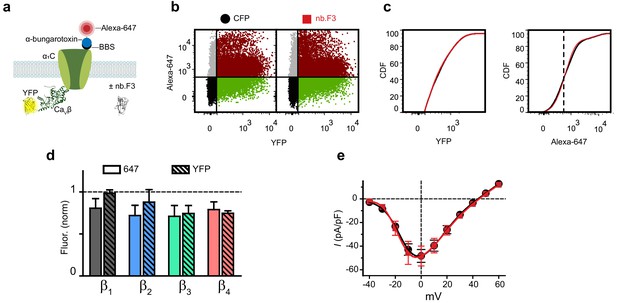
Nb.F3 is functionally silent on reconstituted CaV1.2 channels.
(a) Cartoon of experimental strategy. BBS-α1C was transfected in HEK293 cells with each YFP-CaVβ and either CFP or nb.F3-P2A-CFP. (b) Exemplar flow cytometry dot plot of cells transfected with BBS-α1C, CaVβ1 and CFP (left) or nb.F3 (right). (c) Cumulative distribution histogram of Alexa-647 (left) or YFP fluorescence (right) from CFP (black) or nb.F3 (red) expressing cells. YFP-positive cells (n > 5,000 cells/experiment) were selected for analysis; the threshold for 647 labeling and YFP fluorescence above background is represented with the dashed line. (d) Summary flow cytometry data of surface (647, filled) and total (YFP, patterned) levels of BBS-α1C. Data from nb.F3 was normalized to CFP control group. N = 4 separate experiments, error bars, s.e.m. (e) population I-V curves from whole-cell patch clamp measurements in HEK293 cells expressing α1C, CaVβ2a, and CFP (black, n = 9) or nb.F3 (red, n = 12). Data are means ± s.e.m.
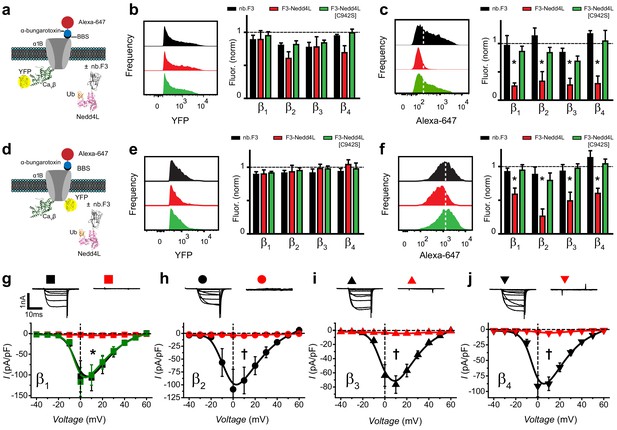
Functional impact of a chimeric nb.F3-Nedd4L protein (CaV-aβlator) on reconstituted Cav2.2 channels.
(a) Schematic of experimental design; HEK293 cells were transfected with BBS-α1B + CaVβ-YFP + α2δ−1, and either nb.F3, nb.F3-Nedd4L, or nb.F3-Nedd4L[C942S]. (b) Exemplar histograms (left) and summary data (right) of flow cytometry experiments measuring total (YFP) levels of CaVβ1b-YFP. Each data set was normalized to a control group that expressed CFP. n > 5,000 cells analyzed per experiment, N = 3 separate experiments, error bars, s.e.m. (c) Exemplar histograms (left) and summary data (right) of flow cytometry experiments measuring surface (647) levels of BBS-α1B. White dashed line is the threshold for 647 signal above background. (d) Experimental strategy; same format as in (a) except YFP was fused to BBS-α1B, enabling measurement of the total levels of the α1B subunit. (e-f) Same format as in (b-d) for cells expressing BBS-α1B-YFP + CaVβ + α2δ-1. (g) Exemplar traces (top) and population I-V curves (bottom) from whole-cell patch clamp measurements in HEK293 cells expressing α1B + CaVβ1b + α2δ-1 and nb.F3 (black, Ipeak, 0mV = -103.5 ± 39.5 pA/pF, n=10), nb.F3-Nedd4L (red, Ipeak, 0mV = -3 ± 0.53 pA/pF, n=11), or nb.F3-Nedd4L[C942S] (green, Ipeak, 0mV = -117 ± 34.8 pA/pF, n=8). (h-j) Same format as (g) for CaV2.2 channels reconstituted with CaVβ2 (h), CaVβ3 (i), and CaVβ4 (j) with nb.F3 (black) or nb.F3-Nedd4L (red). Scale bar 1nA, 10ms. Data are means ± s.e.m., n=10 for each point. *P < 0.05 compared with control, one-way ANOVA with Tukey’s multiple comparison test. †P < 0.01 compared with control, unpaired, two-tailed Student’s t-test.
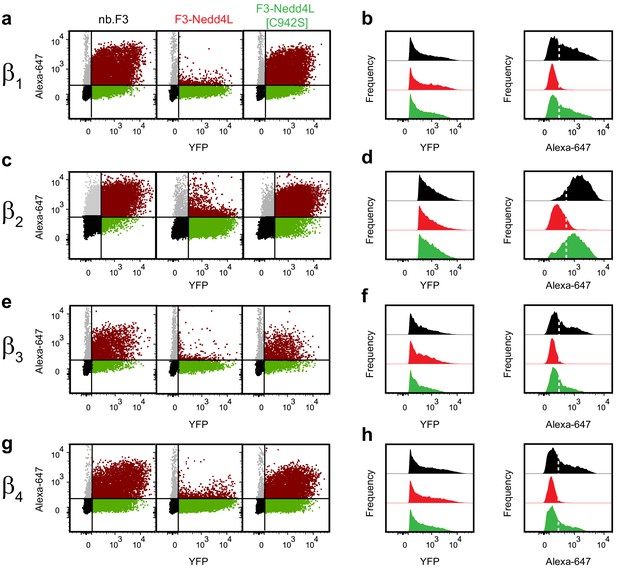
Exemplar flow cytometry data for CaV-aβlation of BBS-α1B with YFP-CaVβ2- CaVβ4.
(a) Exemplar flow cytometry dot plot of cells transfected with BBS-α1B, α2-δ, YFP-CaVβ1 and nb.F3 (left), nb.F3-Nedd4L (middle) or nb.F3-Nedd4L[C942S] (right). (b) Histogram a of YFP (left) or Alexa-647 fluorescence (right) from samples in (a) YFP-positive cells (n > 5,000/experiment) were selected for analysis, the threshold for 647 labeling above background is represented with the dashed line. (c,d) as in (a,b) for cells expressing YFP-CaVβ2. (e,f) as in (a,b) for cells expressing YFP-CaVβ3. (c,d) as in (a,b) for cells expressing YFP-CaVβ2. (g,h) as in (a,b) for cells expressing YFP-CaVβ4.
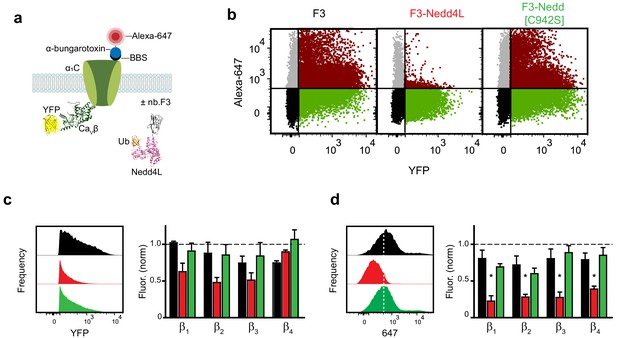
Functional impact of a chimeric nb.F3-Nedd4L protein (CaV-aβlator) on reconstituted Cav1.2 channels.
(a) Schematic of experimental design. HEK293 cells were transfected with BBS-α1C, YFP-CaVβ, and either nb.F3, nb.F3-Nedd4L or nb.F3-Nedd4L[C942S]. (b) Exemplar flow cytometry dot plot of cells transfected with BBS-α1C, YFP-CaVβ1b and nb.F3 (left), nb.F3-Nedd4L (middle) or nb.F3-Nedd4L[C942S] (right). (c,d) Histogram of YFP (c) and Alexa-647 (d) fluorescence from samples in (b) (left) and summary data from N = 3 separate experiments (right). YFP-positive cells (n > 5,000 cells per experiment) were selected for analysis, the threshold for 647 labeling above background is represented with the dashed line. *p<0.05 compared with control, one-way ANOVA with Tukey’s multiple comparison test.
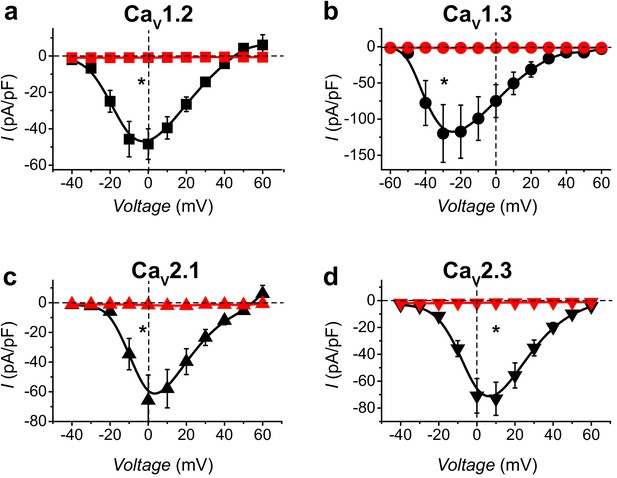
CaV-aβlator inhibits distinct reconstituted HVACCs.
(a) Population I-V curves from HEK293 expressing α1C + β1b + α2δ−1 with either nb.F3 (black, Ipeak, 0mV = −48.4 ± 8.4 pA/pF, n = 12) or CaV-aβlator (red, Ipeak, 0mV = −0.93 ± 0.16 pA/pF, n = 8). (b-d) Same format as (a) for cells expressing reconstituted CaV1.3 (b), CaV2.1 (c), or CaV2.3 (d) channels. Data are means ± s.e.m.†p<0.01 compared with control, unpaired, two-tailed Student’s t-test.
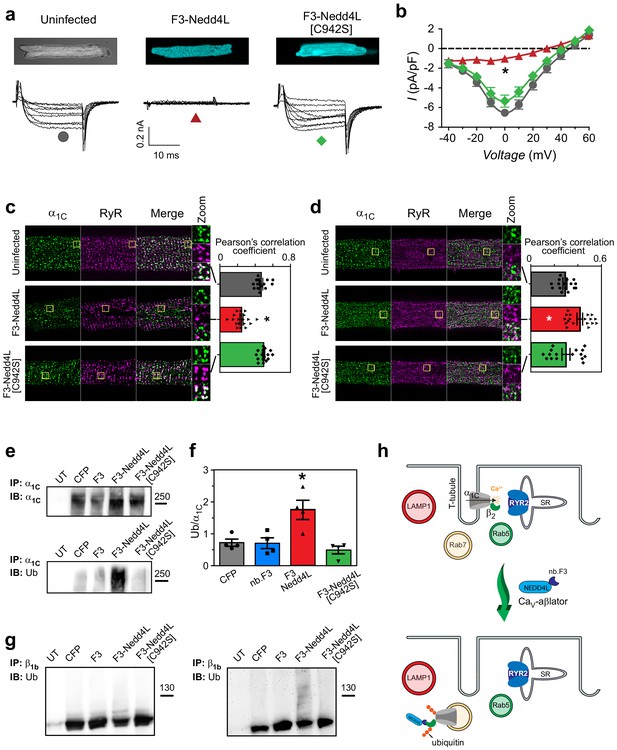
CaV-aβlation of endogenous CaV1.2 in cardiomyocytes.
(a) Confocal images (top) and exemplar traces from whole-cell recordings of uninfected guinea pig cardiomyocytes (left), or infected with adenovirus expressing either CaV-βlator (middle) or nb.F3-Nedd4L[C942S] (right). Scale bar 0.2nA, 10 ms. (b) Population I-V curves from cardiomyocytes expressing CaV-βlator (red), nb.F3-Nedd4L[C942S] (green), or an uninfected control (black). (c) Left, exemplar confocal images of cardiomyocytes fixed and immunostained with α1C (green) and ryanodine receptor (RyR2, magenta) antibodies. Yellow box indicates region of high-zoom merge image.Right, co-localization between α1C and RyR in uninfected cardiomyocytes (gray, PCC = 0.47 ± 0.02, n = 15), and those expressing either CaV-aβlator (red, PCC = 0.24 ± 0.02 n = 19), or nb.F3-Nedd4L[C942S] (green, PCC = 0.50 ± 0.01, n = 17). (d) Left, exemplar confocal images of fixed cardiomyocytes immunostained with α1C (green) and Rab7 (magenta) antibodies. Yellow box indicates region of high-zoom merge image. Right, colocalization between α1C and Rab7 in uninfected cardiomyocytes (gray, PCC = 0.29 ± 0.02, n = 16), and those expressing either CaV-aβlator (red, PCC = 0.42 ± 0.02, n = 18), or nb.F3-Nedd4L[C942S] (green, PCC = 0.30 ± 0.03, n = 16). (e) Pulldown of α1C in HEK293 cells expressing α1C, β1b and either CFP, nb.F3, CaV-aβlator, or nb.F3-Nedd4L-[C942S]. Top, probing pulldown with α1C antibody. Bottom, same blot stripped and re-probed with ubiquitin antibody. (f) Quantification of four separate experiments, as performed in (e). Data are means ± s.e.m for each point. *p<0.05 compared to control, one-way ANOVA with Tukey’s multiple comparison test. (g) Pulldown of CaVβ1b, as in (e). Left, probing with CaVβ1b. Right, same blot stripped and re-probed with ubiquitin antibody. (h) Cartoon illustrating CaV-aβlator-induced relocation of CaV1.2 from dyads to Rab7-positive late endosomes in cardiomyocytes.
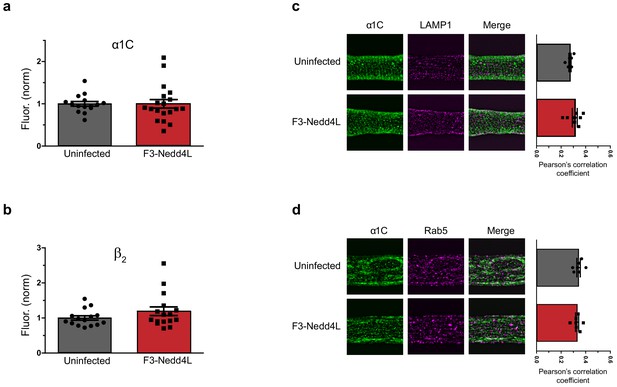
CaV-aβlator does not redistribute α1C to Rab5 early-endosomes or lysosomes, nor decrease total levels of α1C or β2.
(a) Comparison of total α1C levels in uninfected cardiomyocytes (gray, n = 15) and those infected with nb.F3-Nedd4L (red, n = 19). (b). Comparison of total β2 levels in uninfected cardiomyocytes (gray, n = 15) and those infected with nb.F3-Nedd4L (red, n = 16), normalized to uninfected controls. Data are means ± s.e.m. (c) Left, exemplar confocal images of uninfected (top) or F3-Nedd4L-infected (bottom) guinea pig cardiomyocytes, fixed and immunostained with antibodies towards α1C (left) and LAMP1 (middle). Right, colocalization between α1C and Rab5 in uninfected cardiomyocytes (gray, n = 7) and those expressing F3-Nedd4L (red, n = 7). Data are means ± s.e.m. (d) as in (c), showing immunostaining and colocalization analysis of cardiomyocytes immunostained against α1C and Rab5.
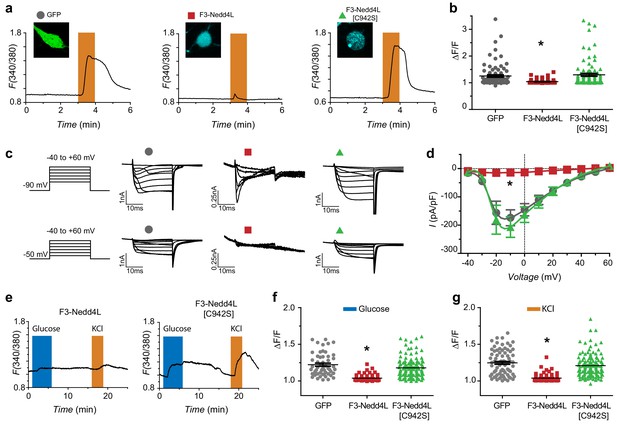
CaV-aβlation of HVACCs in DRG neurons and pancreatic β-cells.
(a) Exemplar Fura-2 traces of murine DRG neurons infected with GFP (left), F3-Nedd4L (middle), F3-Nedd4L[C942S] (right), with confocal images in inset. The orange bars represent depolarization with 40 mM KCl. (b) Summary of maximum responses from neurons infected with GFP (Peak response = 1.25 ± 0.04, n = 84), F3-Nedd4L (1.04 ± 0.01, n = 77), and F3-Nedd4L[C942S] (1.30 ± 0.05, n = 92) in response to 40 mM KCl. Peaks were normalized to the baseline, defined as 1 min prior to the addition of KCl. (c) Exemplar traces of DRG neurons infected with GFP (left), F3-Nedd4L (middle), F3-Nedd4L[C942S] (right). Traces were collected at both a holding potential of −90 mV (top) and −50 mV (bottom). Notably, CaV-aβlator-infected neurons still show robust T-type current when held at −90 mV. (d) Population I-V curves from DRG neurons infected as in (a). Measurements were made at a holding potential of −90 mV. Symbols are mean currents calculated from 15 to 20 ms of a 20 ms test pulse. Data are means ± s.e.m. (e) Exemplar fura-2 traces from dispersed pancreatic islets infected with CaV-aβlator (left) or F3-Nedd4L[C942S] (right) challenged with 16.8 mM glucose (blue bars) and 40 mM KCl (orange bars). (f) Summary of maximum responses from pancreatic β-cells infected with GFP (Peak response = 1.22 ± 0.02, n = 53), F3-Nedd4L (Peak response = 1.04 ± 0.01, n = 62), and F3-Nedd4L[C942S] (Peak response = 1.18 ± 0.01, n = 122) in response to 16.8 mM glucose. (g) Summary of maximum responses from pancreatic β-cells infected with GFP (Peak response = 1.25 ± 0.02, n = 77), F3-Nedd4L (Peak response = 1.04 ± 0.01, n = 62), and F3-Nedd4L[C942S] (Peak response = 1.21 ± 0.01, n = 122) in response to 40 mM KCl. Data are means ± s.e.m. *p<0.05 compared to control, one-way ANOVA with Tukey’s multiple comparison test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (rat) | CACNA1B | NM_147141 | ||
| Gene (rabbit) | CACNA1C | NM_001136522 | ||
| Gene (rat) | CACNB1 | NM_017346 | ||
| Gene (human) | CACNB2 | NM_201590 | ||
| Gene (rat) | CACNB3 | NM_012828.2 | ||
| Gene (rat) | CACNB4 | NM_001105733.1 | ||
| Gene (human) | CACNA2D1 | NM_000722.4 | ||
| Gene (human) | NEDD4L | NM_001144965.2 | ||
| strain, strain background (Escherichia coli) | Rosetta DE3 | Millipore Sigma | ||
| Cell line (Human) | HEK293 | Other | RRID: CVCL_0045 | Laboratory of Dr. Robert Kass |
| Recombinant DNA reagent | nb.F3-CFP-PKCγ | This paper | Made by PCR, see molecular biology and cloning | |
| Recombinant DNA reagent | nb.F3-P2A-CFP | This paper | Made by PCR, see molecular biology and cloning | |
| Recombinant DNA reagent | nb.F3-Nedd4L-P2A-CFP | This paper | Made from pCI HA Nedd4L (Addgene #27000); see molecular biology and cloning | |
| Recombinant DNA reagent | nb.F3-Nedd4L [C942S]-P2A-CFP | This paper | Made by site-directed mutagenesis | |
| Recombinant DNA reagent | BBS-α1B | PMID: 20308247 | ||
| Recombinant DNA reagent | BBS-α1C | PMID: 20308247 | ||
| Antibody | Anti-α1C | Alomone | Cat#: ACC-003 | 1:1000 WB/IF |
| Antibody | Anti-α1C | NeuroMab | Clone: N263/31 | 1:200 IF |
| Antibody | Anti-CaVβ1 | NeuroMab | Clone: N7/18 | 1:500 WB |
| Antibody | Anti-CaVβ2 | Alomone | Cat#: ACC-105 | 1:200 |
| Antibody | Anti-Rab5 | Cell Signaling Technology | Cat#: 3547 | 1:200 |
| Antibody | Anti-Rab7 | Cell Signaling Technology | Cat#: 9367 | 1:200 |
| Antibody | Anti-LAMP1 | Developmental Studies Hybridoma Bank at the University of Iowa | RRID: AB_528127 | 1:100 |
| Antibody | Anti-RyR | Thermo Fisher Scientific | Cat#: MA3-916 | 1:1000 |
| Antibody | Anti-actin | Sigma | Cat#: A5060 | 1:1000 |
| Antibody | Anti-ubiquitin, VU-1 | LifeSensors | Cat#: VU101 | 1:500 |
| Antibody | RFP-trap agarose beads | Chromotek | Cat#: rta-20 | |
| Antibody | Anti-FLAG affinity gel | Sigma-Aldrich | Cat#: A2220 | |
| Peptide, recombinant reagent | FLAG peptide | Sigma-Aldrich | Cat#: F3290 | |
| Peptide, recombinant reagent | Ni-NTA agarose | Qiagen | Cat#: 30210 | |
| Peptide, recombinant reagent | Protein A/G sepharose beads | Rockland | ||
| Peptide, recombinant reagent | α-bungarotoxin, Alexa Fluor 647 conjugate | Life Technologies | ||
| Peptide, recombinant reagent | Fura-2 AM | Life Technologies | Cat#: F1221 | |
| chemical compound, drug | Phorbol 12,13-dibutyrate | Sigma-Aldrich | Cat#: P1269 | |
| commercial assay or kit | AdEasy Adenoviral Vector Systems | Stratagene | ||
| commercial assay or kit | QuikChange Lightning Site-Directed Mutagenesis Kit | Stratagene | ||
| software, algorithm | FlowJo | RRID: SCR_008520 | ||
| software, algorithm | PulseFit | HEKA | ||
| software, algorithm | EasyRatioPro | HORIBA | ||
| software, algorithm | GraphPad Prism | RRID: SCR_002798 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49253.014





