IFI16, a nuclear innate immune DNA sensor, mediates epigenetic silencing of herpesvirus genomes by its association with H3K9 methyltransferases SUV39H1 and GLP
Figures
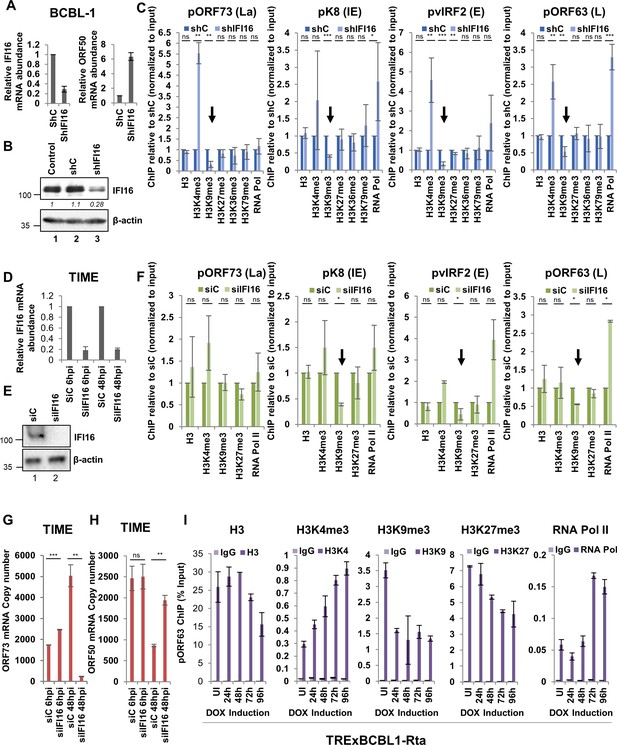
Effect of IFI16 knockdown (KD) on H3K9me3 and RNA Pol II deposition on KSHV lytic gene promoters.
(A) IFI16 was KD in BCBL-1 cells using shRNA lentivirus for 72 hr and KD efficiency was assessed by q-RT PCR and successful induction of lytic KSHV ORF50 gene as a result of IFI16 KD was assessed by q-RT PCR of ORF50. (B) WB showing IFI16 KD compared to untreated or shC treated BCBL-1 cells. (C) ChIP was performed after lentivirus-mediated IFI16 KD in BCBL-1 cells or shC-BCBL-1 cells. Deposition of different histone H3 lysine tri-methylation marks (H3, H3K4me3, H3K9me3, H3K27me3, H3K36me3 and H3K79me3) and RNA Pol II on four different KSHV promoters (pORF73- La, pK8- IE, pvIRF2- E, and pORF63- L) representing the four different temporal KSHV gene classes were tested by q-PCR. ChIP efficiencies normalized to input chromatin are shown as relative to shC control. (D - H) TIME cells were electroporated with either siC or siIFI16. After 72 hr, cells were de novo infected with KSHV (100 DNA copies/cell) for 6 or 48 hr. IFI16 KD efficiencies were assessed by q-RT PCR of the IFI16 gene (D) and WB of IFI16 (E). (F) ChIP was performed after 48 hr of de novo infection of IFI16 KD TIME. (G and H) q-RT PCR (one step TaqMan) of KSHV latent ORF73 (G) and lytic ORF50 (H) mRNA expression normalized to cellular RNaseP after 6 and 48 hr of de novo infection of TIME cells previously treated with siIFI16 of siC for 72 hr. (I) Lytic cycle was induced in TRExBCBL1-RTA cells using doxycycline. At 0, 1, 2, 3 and 4 days post-induction, ChIP was performed. Deposition of different H3 lysine tri-methylation marks (H3, H3K4me3, H3K9me3 and H3K27me3) and RNA Pol II on the ORF63 promoters was tested by q-PCR. ChIP with control IgG was also performed. ChIP efficiencies are represented as % input. Data shown are averages of the results of at least three experiments ± SD. *=p < 0.05; **=p < 0.01; ***=p < 0.001 (unpaired t test).
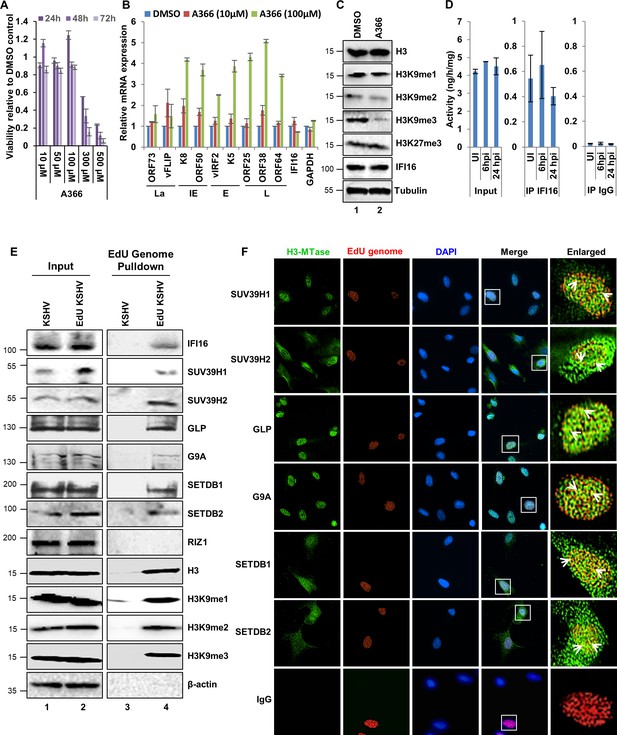
Effect of A366 on KSHV life cycle and the demonstration of IFI16’s association with cellular H3K9 methyltransferase(s) (H3K9 MTase) and recruitment of various H3K9 MTases to the KSHV genome during de novo infection.
(A) MTT cell viability assay of BCBL-1 cells treated with the H3K9me3 specific chemical inhibitor A366 at different concentrations and different time points. (B) q-RT PCR (two-step, sybr Green) of KSHV mRNAs in BCBL-1 cells treated for 72 hr with either vehicle control DMSO or A366 (10 µM and 100 µM). (C) WB of different H3 methylations and IFI16 after A366 treatment of BCBL-1 cells. (D) H3K9 methyltransferase activity (ng/h/mg) assay. TIME cells were infected with KSHV for 6 or 24 hr followed by isolation of nuclear fraction, benzonase treatment and IP with anti-IFI16 or control IgG in the presence of benzonase using the catch and release method. Elution was performed under non-denaturing conditions to keep the associated H3K9 methyltransferase active. H3K9 methyltransferase activity was assayed in the eluate (Materials and methods). *, p<0.05; **, p<0.01; ***,<0.001; unpaired t-test. (E) TIME cells were infected with KSHV genome labeled with EdU or unlabeled control KSHV (100 DNA copies/cell) for 24 hr followed by EdU-KSHV genome pulldown using Click chemistry. The inputs and eluates were blotted for different H3K9 MTases. (F) TIME cells were infected with EdU-labeled KSHV as in (D) and stained using the Click-iT EdU Alexa Fluor 594 Imaging Kit (red). Subsequently, IFA was performed against different H3K9 MTases and colocalization of the IFA signal (green) with KSHV EdU-genome staining (red) resulting in yellow was evaluated (enlarged image, white arrows).
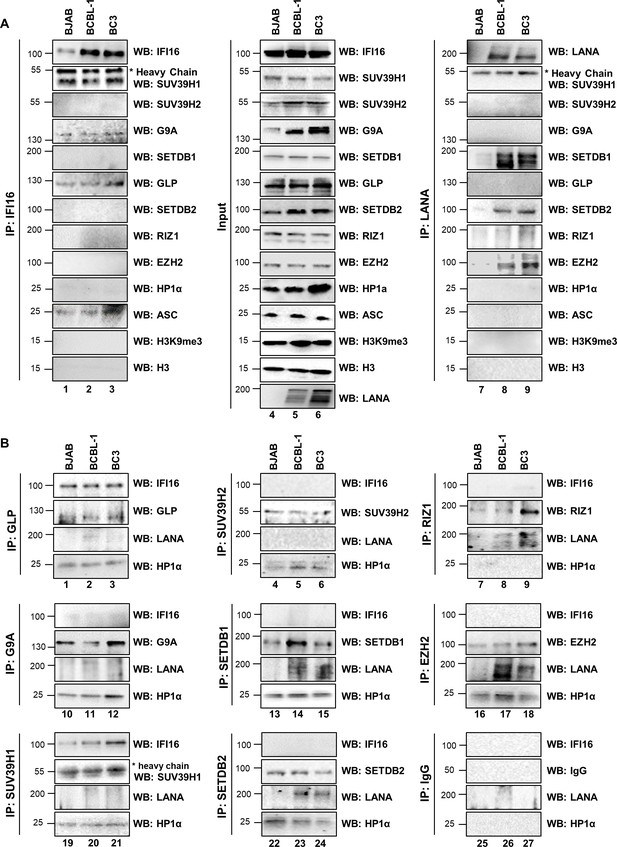
Demonstration of IFI16’s interaction with specific H3K9 MTases in KSHV latently infected PEL (BCBL-1 and BC-3) cells and in uninfected control BJAB cells.
(A) Nuclear fractions were isolated from latently infected cells and uninfected BJAB cells and treated with Benzonase. IPs were performed using anti-IFI16 mAb and LANA mAb and WBs were performed. (B) To confirm IFI16’s and LANA’s interaction with H3K9 MTases, IPs were done with Abs against the H3K9 MTases and blotted for the corresponding MTase, IFI16, LANA and HP1α (heterochromatin protein 1α).
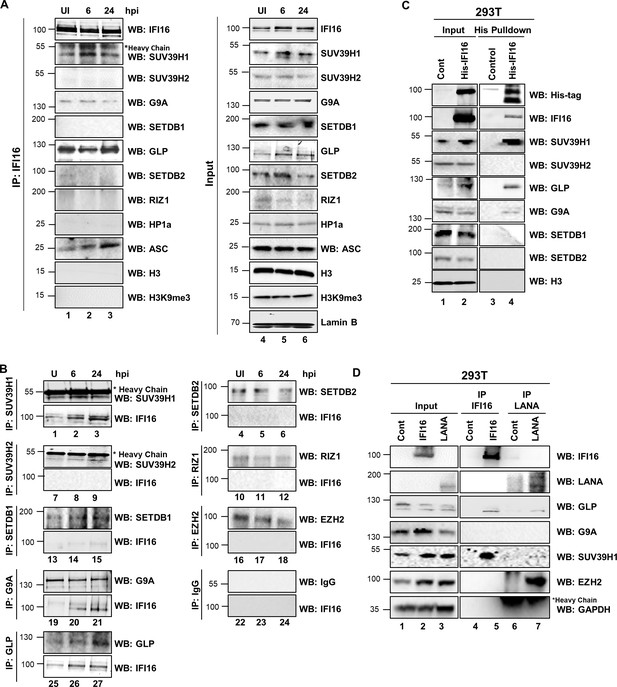
Demonstration of IFI16’s interaction and recruitment of specific H3K9 MTases during de novo KSHV infection.
(A) TIME cells either left uninfected or infected with KSHV for 6 or 24 hr were IPed with anti-IFI16 antibodies and western blotted for the indicated proteins. (B) To confirm IFI16’s interaction with H3K9 MTases, TIME cells were infected as in (A) and IPed with antibodies against the MTases and blotted for the corresponding MTase and IFI16. (C) 293 T cells lacking IFI16 transfected with control plasmid or His-IFI16 expressing plasmid for 72 hr were utilized for His-tag pulldown using HisPur cobalt resin. Inputs and elutions were blotted for the indicated proteins. (D) 293 T cells transfected with control plasmid, IFI16 expressing plasmid or LANA expressing plasmid for 72 hr were IPed with anti-IFI16 mAb or LANA mAb. Inputs and elutions were blotted for the indicated proteins.
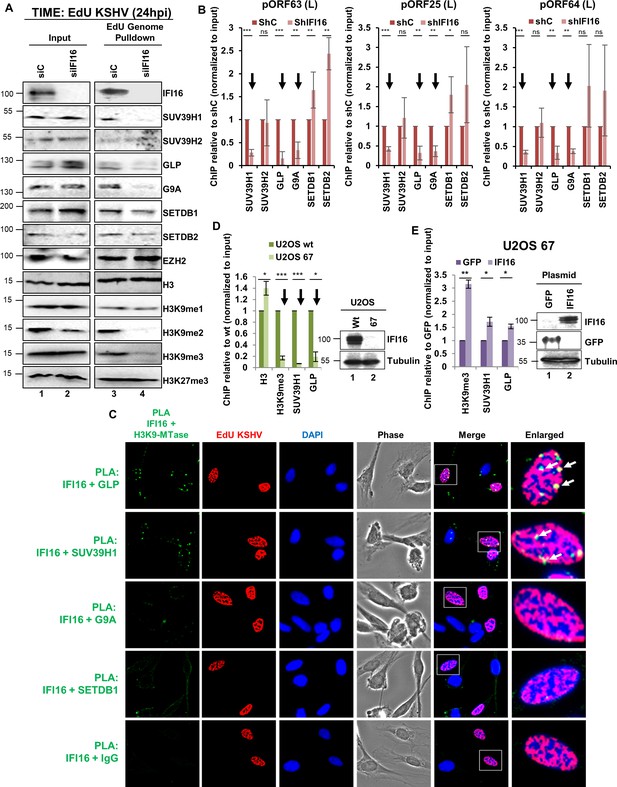
Demonstration of IFI16’s specific interaction with GLP and SUV39H1 and the effect of IFI16 KD upon the recruitment of SUV39H1 and GLP to the KSHV genome during de novo infection.
(A) IFI16 was KD in TIME cells using siRNA and 72 hr later, cells were infected with EdU-KSHV for 24 hr followed by EdU-KSHV genome pulldown using Click chemistry. The inputs and eluates were blotted for different H3K9 MTases and H3K9 methylations. (B) ChIP was performed after shRNA IFI16 KD in BCBL-1 cells and recruitment of different H3K9 MTases on different KSHV promoters (pORF63- L, pORF25- L and pORF64- L) representing promoters where H3K9me3 is most abundantly recruited, were tested by q-PCR. ChIP efficiencies were normalized to input chromatin and are represented as relative to shC control. (C) TIME cells were infected with EdU-KSHV for 24 hr and stained using the Click-iT EdU Alexa Fluor 594 Imaging Kit (red). Subsequently, Proximity Ligation Assay (PLA) was performed to assess the interaction between IFI16 and the indicated H3K9 MTases (green). Colocalization of green (PLA) with red (EdU-KSHV genome) resulting in yellow indicates interaction of IFI16 with the corresponding H3K9 MTases on the KSHV genome (enlarged image, white arrows). Uncropped source PLA data for Figure 5C showing a larger field containing multiple cells are shown in Figure 5—figure supplements 2 and 3. (D) U2OS wt and U2OS 67 (CRISPER Cas-9 IFI16 KO) cells were infected with KSHV (100 DNA copies/cell) for 24 hr, ChIP performed, and the late KSHV promoter pORF63 tested by q-PCR. ChIP efficiencies were normalized to input chromatin and are represented as relative to U2OS wt. WB shows IFI16 KO in U2OS 67 cells. (E) U2OS 67 cells were transfected with either control GFP plasmid or IFI16 plasmid. After 72 hr, cells were infected with KSHV for 24 hr, ChIP performed, and the late KSHV ORF63 promoter was tested by q-PCR. ChIP efficiencies were normalized to input chromatin and are represented as relative to GFP transfected control. The WB shows IFI16 and GFP overexpression in U2OS 67 cells following transfection. Data shown are averages of the results of at least three experiments ± SD (*, p<0.05; **, p<0.01; ***, p<0.001).
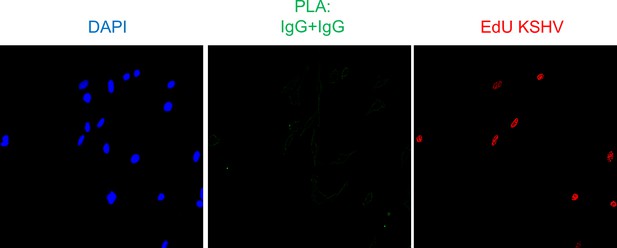
Assessment of the specificity of the PLA reaction.
TIME cells were infected with EdU-KSHV for 24 hr and stained using the Click-iT EdU Alexa Fluor 594 Imaging Kit (red). Subsequently, PLA (green) was performed between IgG (rabbit) and IgG (mouse) to assess the specificity of the PLA reaction.
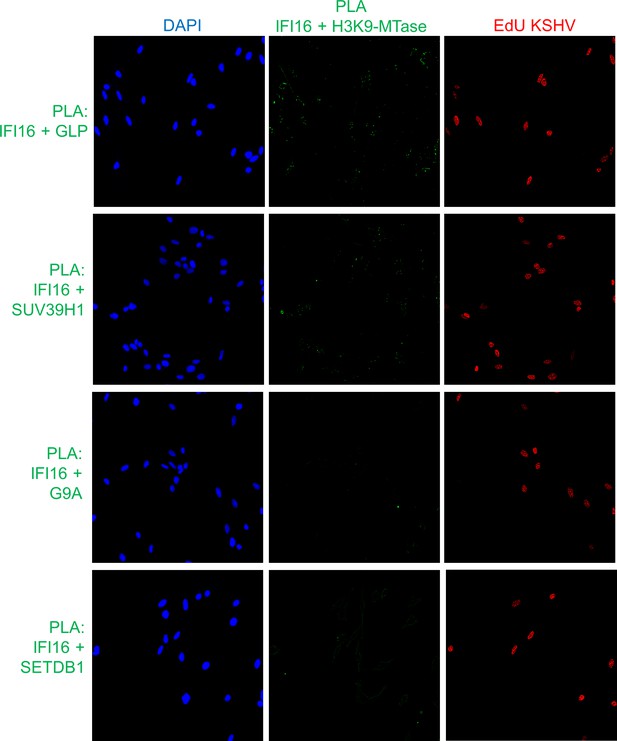
Uncropped source PLA data for Figure 5C.
A larger field containing multiple cells are shown.
Uncropped source PLA data for Figure 5C (continued).
A larger field containing multiple cells are shown.
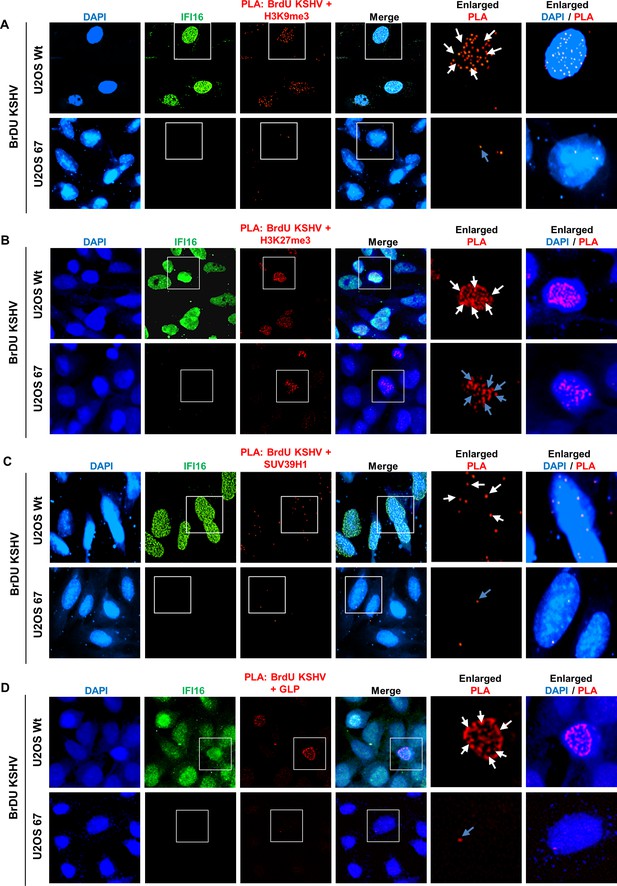
Proximity Ligation Assay (PLA) demonstrating the reduced recruitment of H3K9me3, GLP and SUV39H1 but not H3K27me3 onto the KSHV genome by IFI16 KD.
(A) U2OS wt and U2OS 67 (IFI16 KO) were infected with BrdU genome labeled KSHV for 24 hr and PLA was performed to assess the association between BrdU-KSHV genome DNA and H3K9me3 (red). Following PLA, IFA was performed to stain for IFI16 (green). Colocalization of green (IFA) with red (PLA) resulting in yellow indicates the presence of both, IFI16 and H3K9me3 on the KSHV genome (merged image). In the U2OS 67 panel, as there is no expression of IFI16, yellow colocalization is absent. Boxed areas are enlarged. The number of PLA dots (red) were compared between the U2OS wt (white arrows) and 67 panels (blue arrows). Similar experiments were performed for H3K27me3 (B), GLP(C), and SUV39H1 (D). Uncropped source PLA data for Figure 6 showing a larger field containing multiple cells are shown in Figure 6—figure supplements 2 and 3.
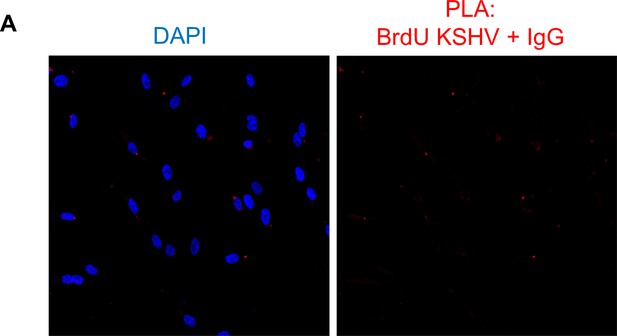
Assessment of the specificity of the PLA reaction.
U2OS wt cells were infected with BrdU genome labeled KSHV for 24 hr and PLA was performed between the BrdU-KSHV genome DNA and IgG (red).
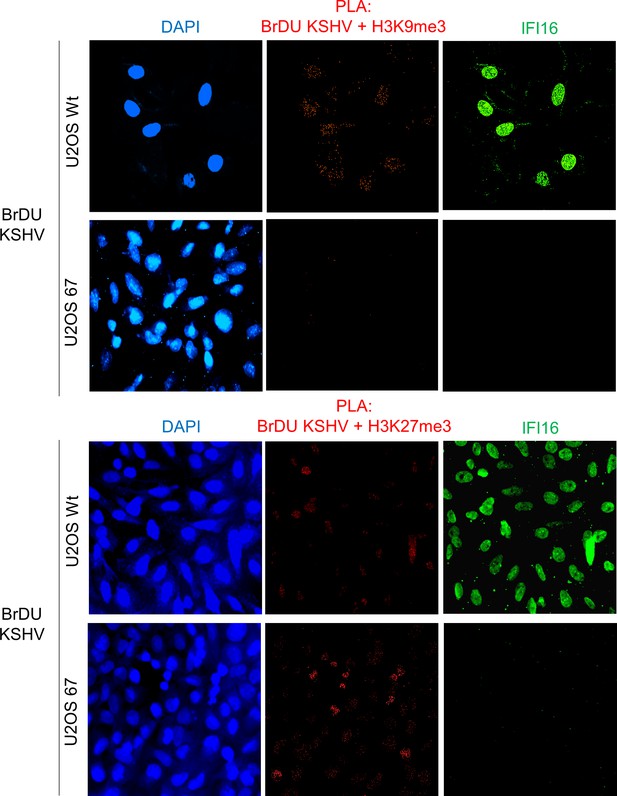
Uncropped source PLA data for Figure 6A and B.
A larger field containing multiple cells are shown.
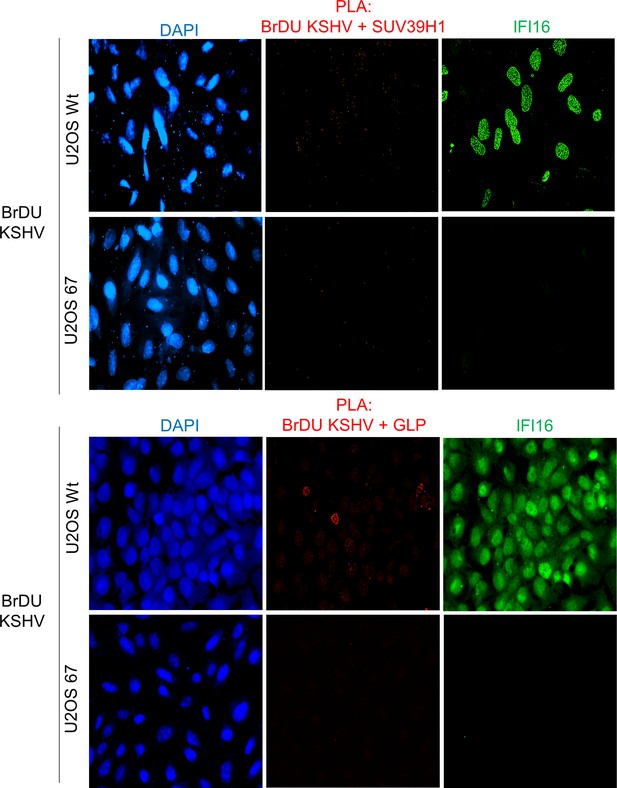
Uncropped source PLA data for Figure 6C and D.
A larger field containing multiple cells are shown.
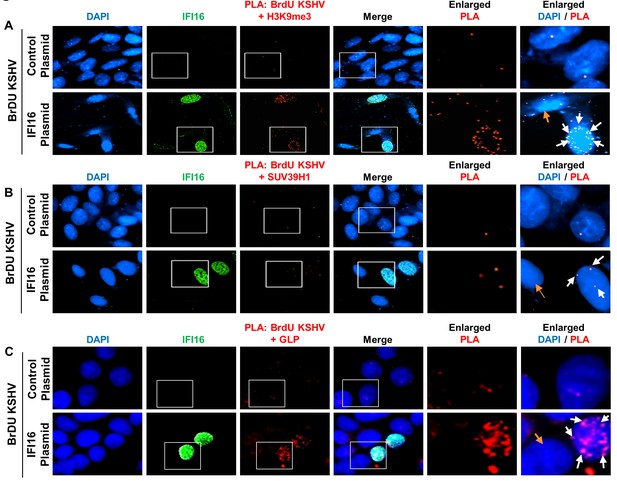
Demonstration of increased recruitment of H3K9me3, GLP and SUV39H1 onto the KSHV genome after IFI16 rescue of IFI16 KO U2OS 67 cells.
(A) U2OS 67 cells were transfected with control plasmid or IFI16 expressing plasmid. After 72 hr, cells were infected with BrdU-KSHV for 24 hr. Following infection, PLA was performed to assess the interaction between the BrdU-KSHV genome DNA and H3K9me3 (red). Subsequently, IFA was performed to stain for IFI16 (green). The number of PLA dots (red) can be compared between the different panels. Boxed areas are enlarged. The white arrows show PLA dots in a cell expressing transfected IFI16, while the orange arrow show a cell that has not been transfected with IFI16 in the same field. A similar experiment was performed for SUV39H1 (B) and GLP (C). Uncropped source PLA data for Figure 7 showing a larger field containing multiple cells are shown in Figure 7—figure supplements 2 and 3.
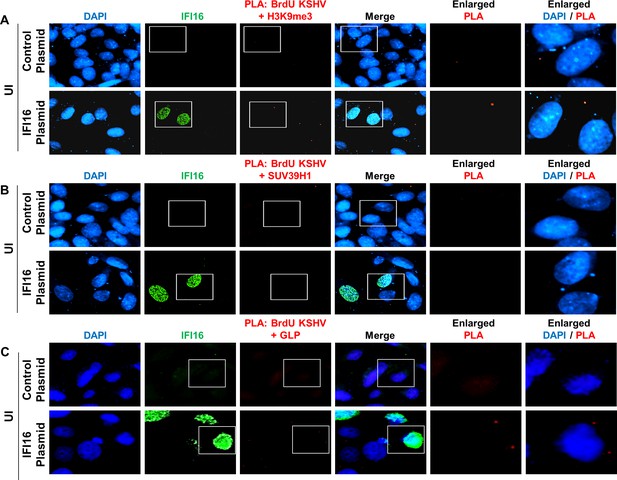
Assessment of the specificity of the PLA reaction.
(A) [Supplementary to Figure 7] U2OS 67 cells were transfected with control plasmid or IFI16 expressing plasmid. After 72 hr, cells were mock infected (UI) for 24 hr. Following infection, PLA was performed to assess the specificity of the PLA reaction. Subsequently, IFA was performed to stain for IFI16 (green). The number of PLA dots (red) can be compared between the different panels. Boxed areas are enlarged. Specificity of the PLA reaction can be ascertained by the lack of PLA dots in the mock-infected cells. Similar experiments were performed for SUV39H1 (B) and GLP (C).
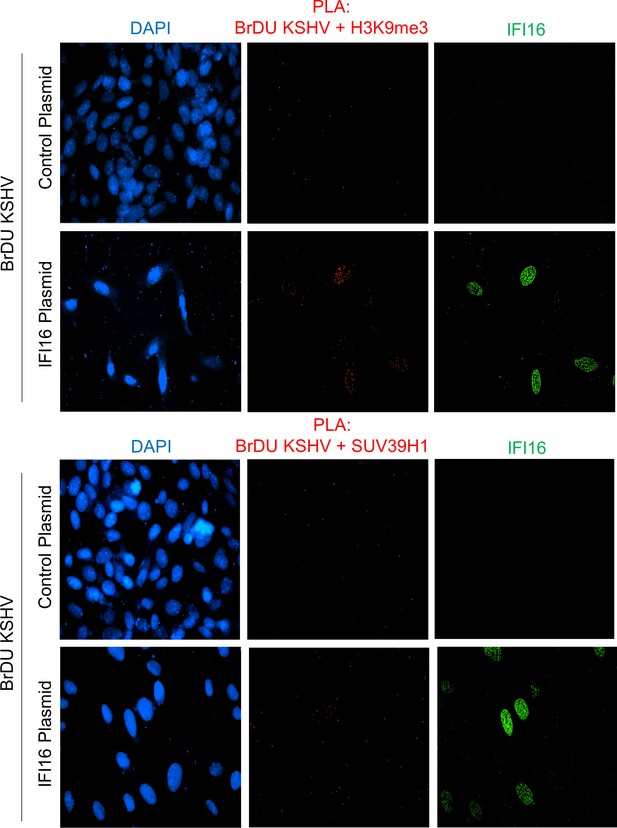
Uncropped source PLA data for Figure 7A and B.
A larger field containing multiple cells are shown.
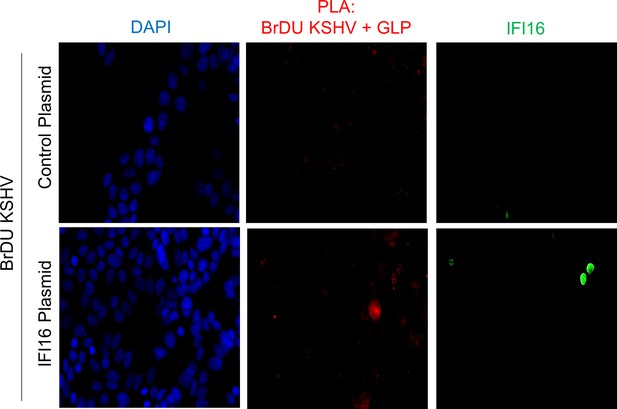
Uncropped source PLA data for Figure 7C.
A larger field containing multiple cells are shown.
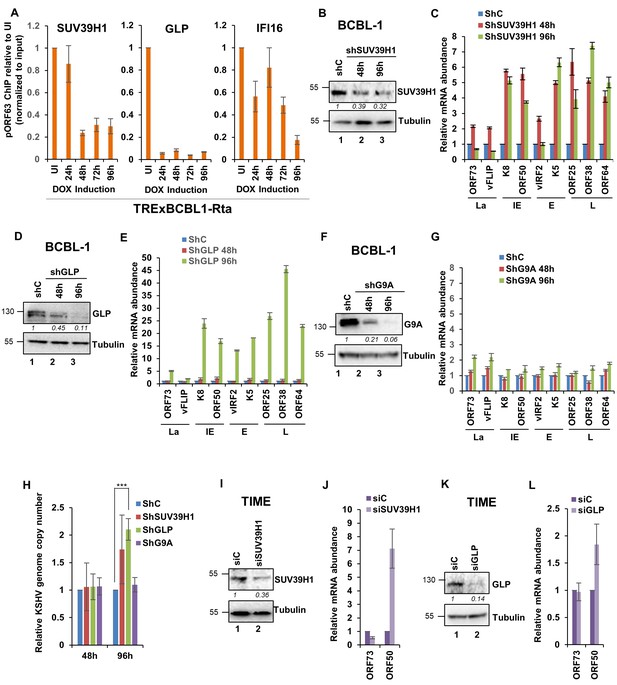
Demonstration of the essential role of H3K9MTAses SUV39H1, and GLP but not G9A in the regulation of KSHV genes during latency and de novo infection.
(A) TRExBCBL1-RTA cells were induced with doxycycline and at 0, 1, 2, 3 and 4 days post-induction, ChIP was performed against the indicated proteins. Deposition of GLP, SUV39H1 and IFI16 on the ORF63 promoter was tested by q-PCR. ChIP efficiencies normalized to input chromatin are shown as relative to uninduced (UI) control. (B) SUV39H1 was knocked down in BCBL-1 cells using lentivirus shRNA for 72 hr and KD efficiency was assessed by WB. (C) mRNA levels of KSHV genes were assessed by real-time RT-PCR after SUV39H1 KD. (D) GLP was knocked down in BCBL-1 cells using lentivirus shRNA for 72 hr and KD efficiency assessed by WB. (E) mRNA levels of KSHV genes were assessed by real-time RT-PCR after GLP KD. (F) G9A was knocked down in BCBL-1 cells using lentivirus shRNA for 72 hr and KD efficiency assessed by WB. (G) mRNA levels of KSHV genes were assessed by real-time RT-PCR after G9A KD. All mRNA levels were normalized against β-tubulin mRNA and are expressed as relative amounts compared to shC treatments. (I) TIME cells were electroporated with siC or a SUV39H1-specific siRNA pool. After 72 hr, cells were de novo infected with KSHV DNA copies/cell) for 48 hr. SUV39H1 KD efficiency was assessed by WB. (H) Real-time DNA-PCR showing the KSHV genome copy numbers at 2 and 4 days post-KD of SUV39H1, GLP and G9A in BCBL-1 cells. Primers specific to the ORF73 gene were used and the level of genomic DNA was normalized against the β-tubulin gene. (J) mRNA expression of ORF73 and ORF50 genes was evaluated using the TaqMan method, normalized to cellular RNaseP and are expressed as relative to siC treatment. (K) GLP KD efficiency was assessed by WB in TIME cells that were electroporated with siC or a GLP-specific siRNA pool. After 72 hr, cells were de novo infected with KSHV for 48 hr. (L) mRNA expression of ORF73 and ORF50 genes was evaluated by TaqMan method, normalized to cellular RNaseP and are expressed as relative to siC treatment. Data shown are averages of the results of at least two experiments ± SD *, p<0.05; **, p<0.01; ***, p<0.001.
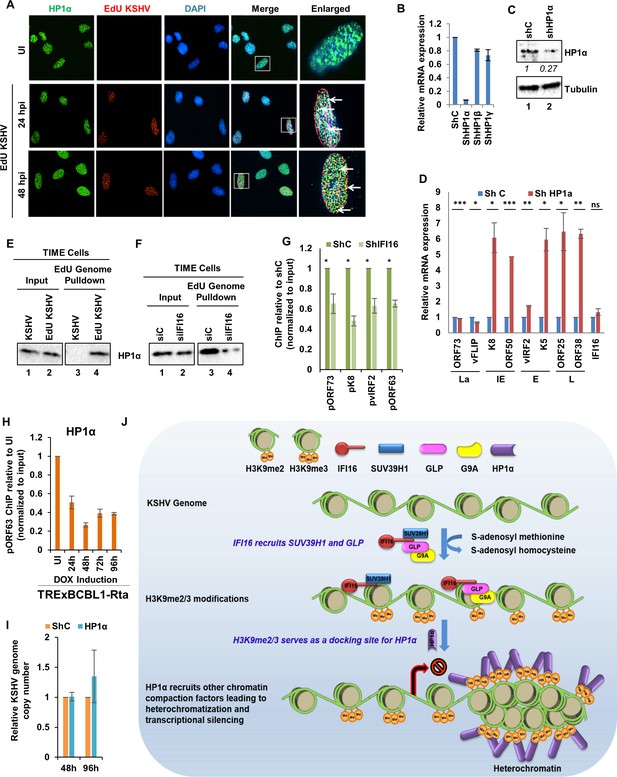
Demonstration of IFI16 mediated H3K9me3 dependent recruitment of Heterochromatin Protein 1-α (HP1α).
(A) TIME cells were infected with EdU-KSHV and stained using the Click-iT EdU Alexa Fluor 594 Imaging Kit (red). Subsequently, IFA was performed against HP1α (green). Colocalization of green (IFA) with red (EdU-KSHV genome) resulting in yellow indicates recruitment of HP1α on the KSHV genome (enlarged image, white arrows). (B) HP1α was KD in BCBL-1 cells by shRNA for 96 hr and KD efficiency assessed by qRT PCR using primers specific for HP1α as well as HP1β and HP1γ. (C) WB to confirm efficient KD. (D) KSHV mRNA levels were assessed by qRT PCR after HP1α KD. IFI16 mRNA was also assessed to confirm that the effect is specific for the KSHV genome only. mRNA levels were normalized against β-tubulin mRNA and expressed as relative amounts compared to shC-treated cells. (E) TIME cells were either infected with EdU-KSHV or control KSHV for 24 hr followed by EdU-KSHV genome pulldown using Click chemistry. The inputs and eluates were blotted for the presence of HP1α. (F) IFI16 was KD in TIME cells using siIFI16. After 72 hr, cells were infected with EdU-KSHV for 24 hr followed by EdU-KSHV genome pulldown using Click chemistry. The inputs and eluates were blotted for the presence of HP1α. (G) HP1α ChIP was performed 48 hr of de novo infection of TIME cells previously KD of IFI16 for 72 hr. KSHV promoters pORF73- La, pK8- IE, pvIRF2- E, and pORF63- L were tested by q-PCR. ChIP efficiencies have been normalized to input chromatin and are represented as relative to shC control. Data shown are averages of the results of at least three experiments ± SD (*, p<0.05; **, p<0.01; ***, p<0.001. (H) TREXBCBL1-RTA cells were induced with doxycycline and at 0, 1, 2, 3 and 4 days post-induction, ChIP was performed against HP1α. Deposition of HP1α on the ORF63 promoter was tested by q-PCR. ChIP efficiencies normalized to input chromatin are shown as relative to uninduced (UI) control. (I) Real-time DNA PCR showing KSHV genome copy number 2 and 4 days post KD of HP1α in BCBL-1 cells. Primers specific to the ORF73 gene were used and the level of genomic DNA was normalized against the β-tubulin gene. (J) Schematic model showing the role of IFI16 in recruiting and maintaining H3K9 MTase SUV39H1 and GLP onto the KSHV genome leading to tri-methylation (me3) of H3K9. Establishment of H3K9me3 marks on the KSHV genome leads to the recruitment of heterochromatin protein HP1α which in turn leads to the DNA compaction and transcription silencing.
Tables
List of antibodies used and their dilutions for different applications.
https://doi.org/10.7554/eLife.49500.020| Antibody | Raised in | Company/catalogue# | Dilution used |
|---|---|---|---|
| IFI16 (1G7) | mouse monoclonal | Santa Cruz Biotechnology (#SC-8023) | WB: 1:700 IP: 1:30 ChIP: 1:30 IFA: 1:100 PLA: 1:50 |
| LANA | Rabbit polyclonal | Raised in-house (Uk183) | WB: 1:1000 IP: 1:50 |
| H3 (D2B12) | Rabbit monoclonal | Cell Signaling Technology (#4620S) | WB: 1:5000 ChIP: 1:50 |
| H3K9me1 | Rabbit polyclonal | Abcam (#ab8896) | WB: 1:1000 |
| H3K9me2 [mAbcam 1220] | Mouse monoclonal | Abcam (#ab1220) | WB: 1:1000 |
| H3K9me3 | Rabbit polyclonal | Active Motif (#39161) | WB: 1:2000 IP: 1:50 ChIP: 1:50 PLA: 1:100 |
| H3K27me3 | Rabbit polyclonal | Active Motif (#39155) | WB: 1:2000 IP: 1:50 ChIP: 1:50 PLA: 1:100 |
| H3K79me3 | Rabbit polyclonal | Abcam (#ab2621) | ChIP: 1:30 |
| H3K4me3 | Rabbit polyclonal | Active Motif (#39159) | WB: 1:2000 IP: 1:50 ChIP: 1:50 PLA: 1:100 |
| H3K36me3 | Rabbit polyclonal | Abcam (#ab9050) | ChIP: 1:30 |
| RNA Pol II CTD [8WG16] | Mouse monoclonal | Abcam (#ab817) | ChIP: 1:50 |
| SUV39H1 (MG44) | Mouse monoclonal | Active Motif (# 39785) | WB: 1:500 IP: 1:30 ChIP: 1:30 IFA: 1:100 PLA: 1:50 |
| SUV39H2 | Goat polyclonal | Novus Biologicals (#NB-100–1140) | WB: 1:500 IP: 1:30 ChIP: 1:30 IFA: 1:100 |
| GLP (B0422) | Mouse monoclonal | Novus Biologicals (# PPB0422-00) | WB: 1:1000 IP: 1:50 ChIP: 1:50 IFA: 1:100 PLA: 1:100 |
| G9A (C-9) | Mouse monoclonal | Santa Cruz Biotechnology (#sc-515726) | WB: 1:500 IP: 1:30 ChIP: 1:30 IFA: 1:70 PLA: 1:50 |
| SETDB1 | Rabbit polyclonal | Novus Biologicals (#NBP2-20322) | WB: 1:500 IP: 1:30 ChIP: 1:30 IFA: 1:70 PLA: 1:50 |
| SETDB2 | Goat polyclonal | Novus Biologicals (#NB100-1137) | WB: 1:500 IP: 1:30 ChIP: 1:30 IFA: 1:70 |
| RIZ1 (N-terminal) | Rabbit polyclonal | Abcam (#ab198792) | WB: 1:500 IP: 1:30 |
| HP1α (GA-62) | Rabbit polyclonal | Cell Signaling Technology (#2616S) | WB: 1:1000 IP: 1:50 ChIP: 1:50 IFA: 1:100 PLA: 1:100 |
| EZH2 (EPR20108) | Rabbit monoclonal | Abcam (#191250) | WB: 1:5000 IP: 1:100 |
| ASC (TMS1) | Mouse monoclonal | MBL International Corporation (#D086-3) | WB: 1:1000 |
| His-tag (HIS.H8) | Mouse monoclonal | Thermo Fisher Scientific (#MA1-21315)) | WB: 1:5000 |
| β-Tubulin (D66) | Mouse Monoclonal | Sigma-Aldrich (#T0198) | WB: 1:5000 |
| β Actin (AC15) | Mouse monoclonal | Sigma-Aldrich (#A5441) | WB: 1:5000 |
| GAPDH | Rabbit polyclonal | Proteintech (#10494–1-AP) | WB: 1:5000 |
| GFP (GF28R) | Mouse monoclonal | Thermo Fisher Scientific (#MA5-15256) | WB: 1:4000 |
| Lamin B | Rabbit polyclonal | Abcam (#ab16048) | WB: 1:1000 |
| Normal IgG Rabbit | Rabbit | Cell Signaling Technology (#2729) | WB: 1:1000 IP: 1:50 ChIP: 1:50 IFA: 1:100 |
| Mouse IgG2a (MOPC-173) | Mouse | Abcam (#ab18413) | WB: 1:1000 IP: 1:50 ChIP: 1:50 IFA: 1:100 |
| BrdU | Rabbit polyclonal | Rockland antibodies and assays (#600–401 C29) | PLA: 1:100 |
| BrdU (MoBU-1) | Mouse monoclonal | Thermo Fisher Scientific (#B35128) | PLA: 1:100 |
| Anti-mouse HRP | Sheep polyclonal | GE Healthcare (#NA931V) | WB: 1:5000 |
| Anti-rabbit HRP | Donkey polyclonal | GE Healthcare (#NA934V) | WB: 1:5000 |
| Anti-goat HRP | Mouse | Santa Cruz Biotechnology (#sc-2354) | WB: 1:5000 |
| Anti-rabbit IgG Conformation Specific HRP (L27A9) | Mouse monoclonal | Cell Signaling Technology (#5127S) | WB: 1:500 |
| Anti-mouse Light Chain specific HRP | Goat polyclonal | Jackson ImmunoResearch (#115–035174) | WB: 1:1000 |
| Anti-rabbit Light Chain specific HRP | Mouse monoclonal | Jackson ImmunoResearch (#211-032-171) | WB: 1:1000 |
| Anti-mouse IgG (H+L)-Alexa fluor 488 | Goat polyclonal | Thermo Fisher Scientific (#A11029) | IFA: 1:500 |
| Anti-rabbit IgG (H+L)-Alexa fluor 488 | Donkey polyclonal | Thermo Fisher Scientific (#A21206) | IFA: 1:500 |
| Anti-goat IgG (H+L)-Alexa fluor 488 | Donkey polyclonal | Thermo Fisher Scientific (#A11055) | IFA: 1:500 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.49500.021





